Figure 5: MPRAu uncovers functional sequence architectures via SNV and deletion tiling.
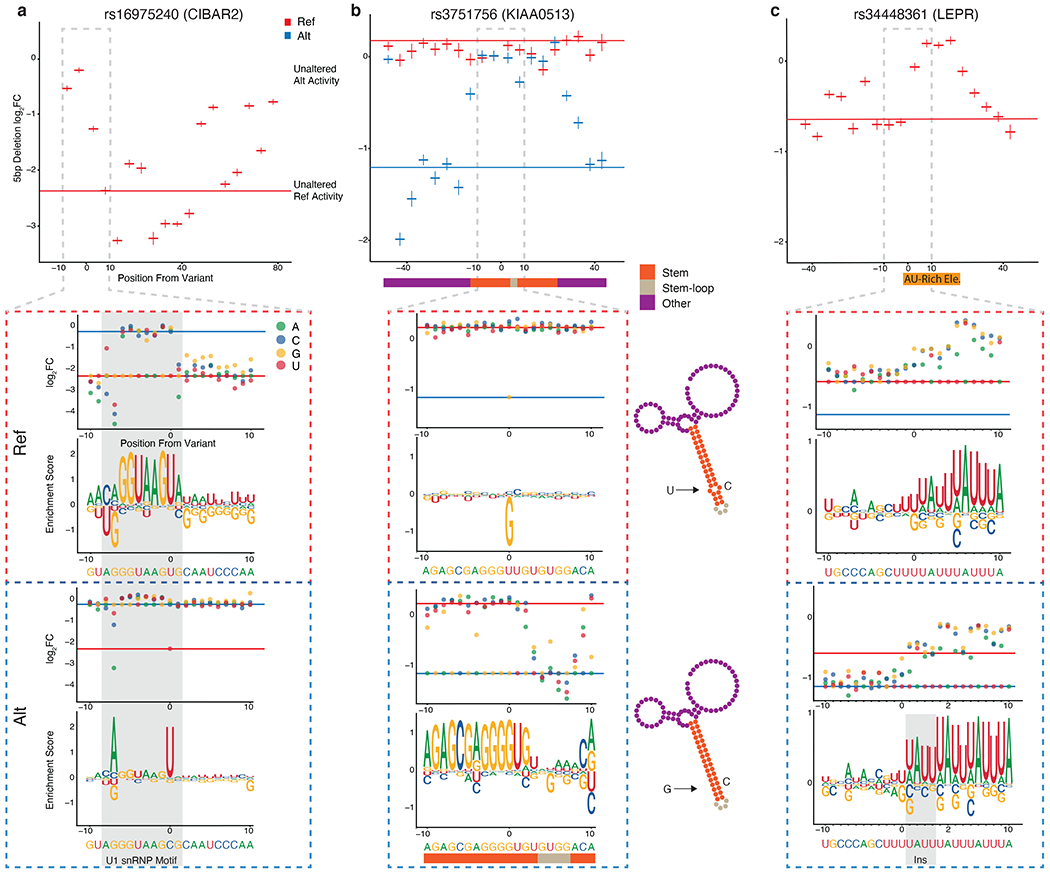
For a-c, Top, 5 bp deletion tiling of the targeted variant using the ref (red) or alt (blue) sequence context across 100 bp. Bottom, SNV tiling +/− 10 bp around each variant showing log2 fold change and motif enrichment score in both variant contexts. HEK293 3’UTR activity data is plotted for all panels. a, Plots for tiling at rs16975420 (CIBAR2), including the sequence motif for U1 snRNP (gray shading). b, Plots for tiling at rs3751756 (KIAA0513), overlaid with a predicted structure of the RNA element (bottom middle). Large magnitude changes observed in SNV tiling are predicted to disrupt bases in the stem structure. c, Plots for tiling at rs34448361 (LEPR) with an AU-rich element denoted.
