Figure 6: rs705866 and rs1059273 are endogenously validated tamVars impacting human disease and adaptation phenotypes.
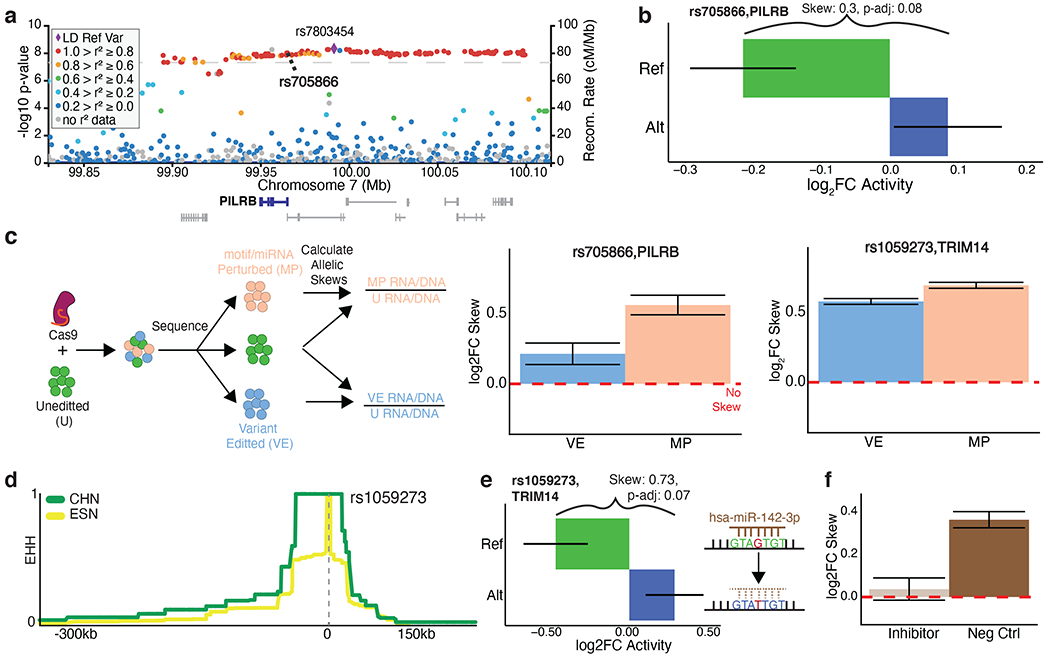
a, Age-related macular degeneration GWAS association plot surrounding the tag SNP rs7803454 with rs705866 (MPRAu tested SNP) in bold. b, MPRAu allelic results for rs705866. c, Schematic of the HDR experimental pipeline (left). Allelic skews estimated from HDR for rs705866 (center), and rs1059273 (right) exhibit the same directionality as the MPRAu results. d, EHH score surrounding rs1059273 suggests evolutionary selection in Han Chinese (CHN). For comparison, EHH scores for Esan in Nigeria (ESN) are also shown. e, MPRAu shows significant attenuating activity in the allelic background (ref) with the unperturbed miRNA binding site for rs1059273. f, Allelic skew result after transfection of a miRNA inhibitor for hsa-miR-142-3p versus a negative control miRNA inhibitor.
