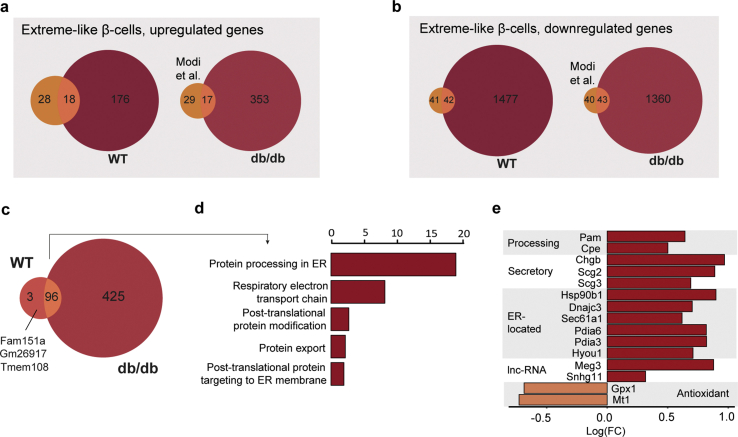
FigS5.
Transcriptional characterization of extreme-like β-cells in T2D. a–b, Comparison of (a) upregulated and (b) downregulated genes from the Ins2-high β-cell population described by Modi et al. (2019) with db/db extreme-like β-cells and WT extreme-like β-cells from our dataset, thresholded at estimated logFC > 0.2 or logFC < –0.2, B > 50, p(adj) < 0.01. c, Venn diagram showing the overlap of enriched genes of WT- and db/db-derived (Sham, PF, VSG) extreme-like β-cells. Limma-trend method was used to find differentially expressed genes at absolute log(fold change) > 0.15 and B > 100. d, GO and KEGG pathway enrichment analysis using EnrichR webtool with Fisher's exact test revealed significantly upregulated ontologies for genes commonly overexpressed in both WT and db/db extreme-like β-cells. Venn diagrams aided in identifying the overlapping genes that were group invariant. X-axis indicates –log10(adjusted p-value). e, Bar plot showing selected regulated genes in db/db-derived (Sham, PF, VSG) extreme-like β-cells.