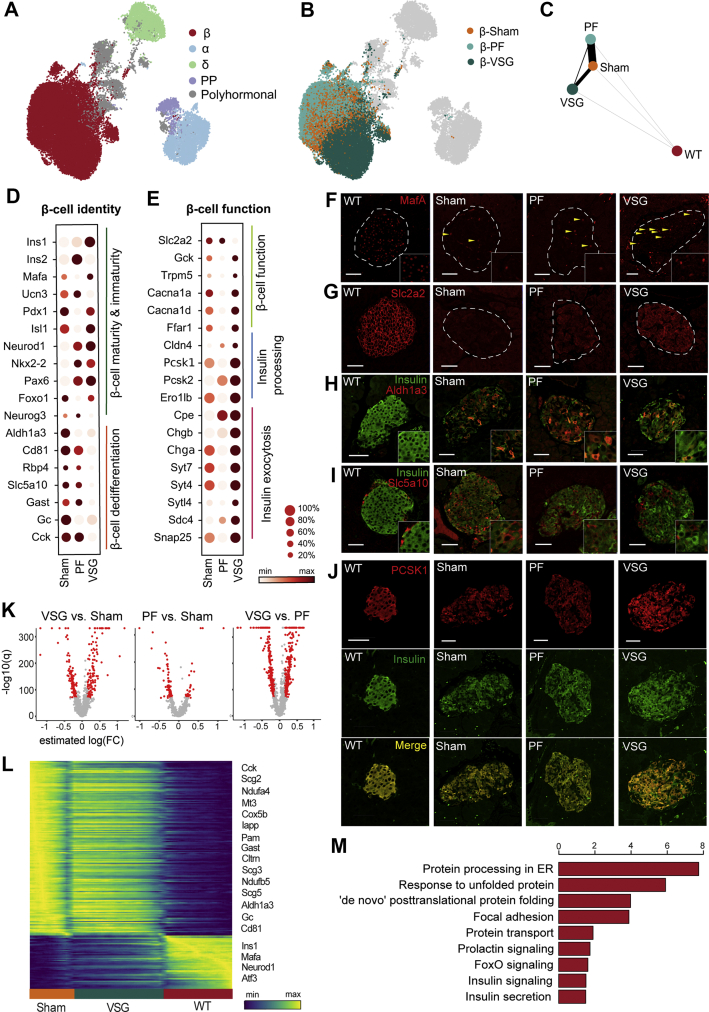
Figure 3.
VSG distinctly improves the identity and function of β-cells. A,B, UMAP plots of 35,941 endocrine cells from Sham, VSG and PF. Colours in (a) indicate the five main endocrine cell clusters and in (b) distinguish β-cells specifically according to the three groups. C, Abstracted graph of cluster connectivity assessing transcriptomic similarity between VSG-, PF-, Sham- and WT-derived β-cells, inferred using PAGA. PAGA is based on a statistical method that measures relatedness between single-cell clusters, with edge weights indicating link significance. D,E, Gene expression for key genes involved in β-cell identity and function changed upon VSG. (d) Dot plot showing that VSG-derived β-cells upregulated essential maturity markers and downregulated dedifferentiation markers. (e) Dot plot showing genes critical for β-cell function and metabolism, insulin processing and exocytosis. Color intensity shows normalized mean expression in a group; dot size indicates the percentage of cells that express the corresponding gene. Expression is scaled per gene. F–J, Immunohistochemical analysis of MafA, Slc2a2, Insulin, Aldh1a3, Slc5a10 and Pcsk1 in β-cells of WT, Sham, PF and VSG mice at study end. Scale is 50 μm K, Volcano plots indicating differences in the number of genes that were differentially expressed when comparing β-cells from Sham and VSG versus β-cells from Sham and PF, obtained from differential analysis testing using limma-trend. Red dots denote significantly regulated genes thresholded on significance levels (absolute estimated log (fold change (FC))>0.15 and B > 150). p-values were corrected for multiple testing using BH. L, Heatmap showing expression changes in genes that were differentially regulated across Sham, VSG and WT β-cells. As a first step, a trajectory of β-cell dedifferentiation upon treatment was estimated using diffusion pseudotime (DPT) and randomly assigning a root cell within the dedifferentiated cells. β-cells were further grouped according to Sham, VSG and WT and gene expression trends were visualized along this one-dimensional axis. Y-axis shows normalised gene expression as the running average along the inferred trajectory, scaled to the maximum observed level per gene. M, Enrichment analysis of GO terms and KEGG pathways of upregulated genes at estimated log (FC) > 0.15 and B > 150 in β-cells between VSG and PF. Method limma-trend was used to identify differentially expressed genes and gene-set enrichment was done with EnrichR webtool using Fisher's exact test to find enriched terms and pathways. X-axis indicates –log10 (adjusted p-value).