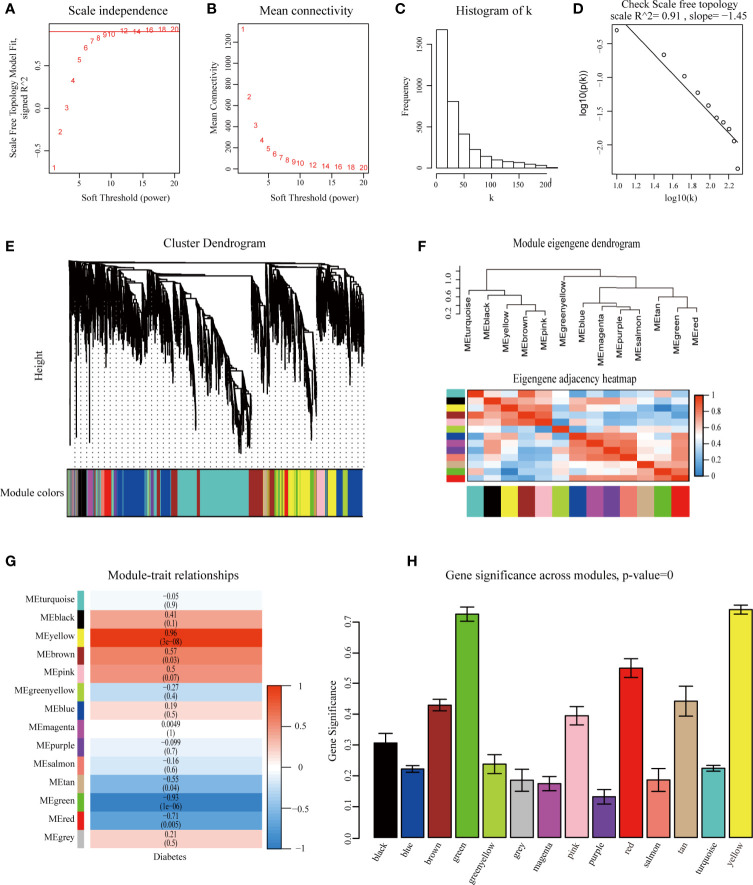
Figure 2.
Identification of modules associated with diabetic cardiomyopathy using the weighted gene co-expression network analysis (WGCNA). (A) Analysis of the scale-free fit index for various soft-thresholding powers (β). (B) Analysis of the mean connectivity for various soft-thresholding powers. (C) Histogram of connectivity distribution when β=12. (D) Checking the scale-free topology when β=12. Identification of modules associated with diabetic cardiomyopathy. (E) Dendrogram of the top 25% most variant genes clustered based on a dissimilarity measure (1-TOM). (F) Module eigengene dendrogram and eigengene network heatmap summarize the modules yielded in the clustering analysis. (G) Heatmap of the correlation between module eigengenes and diabetic cardiomyopathy. (H) Distribution of average gene significance and errors in the modules associated with diabetic cardiomyopathy. TOM, Topological Overlap Matrix.