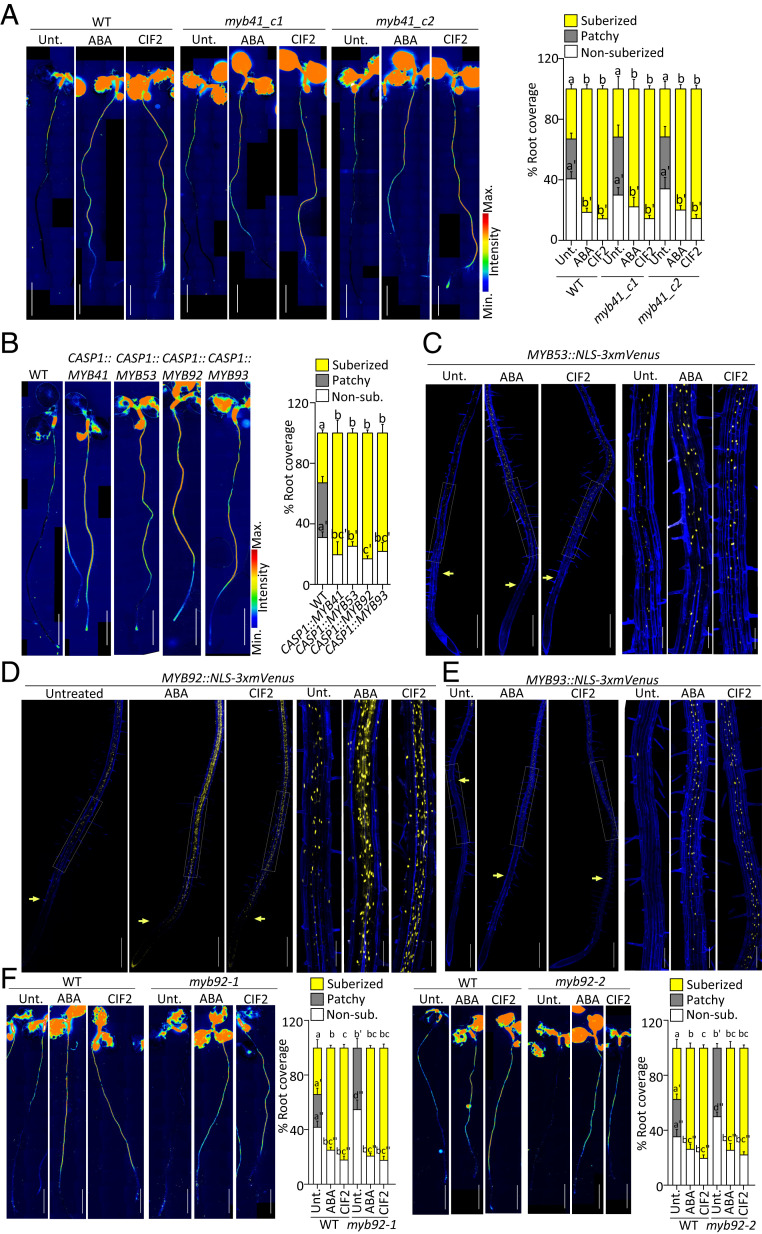
Fig. 3.
MYB53, MYB92, and MYB93 regulation and function in suberin induction. (A, B, and F) FY staining for suberin. Whole-mount staining (Left) and quantifications of suberin pattern along the root (Right), n ≥ 10, error bars, SD. (Scale bars, 2 mm.) (A) FY staining of WT and two myb41 CRISPR mutant alleles, myb41_c1 and myb41_c2 untreated or treated with 1 µM ABA or 1 µM CIF2 for 16 h. Different letters indicate significant differences between conditions for a given genotype (P < 0.05). (B) FY staining of WT, CASP1::MYB41, CASP1::MYB53, CASP1::MYB92, and CASP1::MYB93 lines. Different letters indicate significant differences between conditions (P < 0.05). (C) MYB53::NLS-3xmVenus, (D) MYB92::NLS-3xmVenus, and (E) MYB93::NLS-3xmVenus expression (in yellow) untreated or after treatments with 1 µM ABA or 1 µM CIF2 for 16 h. (C–E) Pictures are presented as maximum intensity Z projections taken from the root tip to 4 to 5 mm (Left) with zoomed views corresponding to the zone of patchy suberization in untreated plants (Right). Arrows highlight the onset of expression. Propidium iodide (PI, in blue) was used to highlighted cells. (Scale bars, 500 μm [Left] and 125 μm [Right].) Quantifications are shown in SI Appendix, Fig. S3 D–F. (F) FY staining of WT, and myb92 mutants untreated or treated with 1 μM ABA or 1 μM CIF2 for 16 h. Different letters indicate significant differences between conditions for a given genotype (P < 0.05).