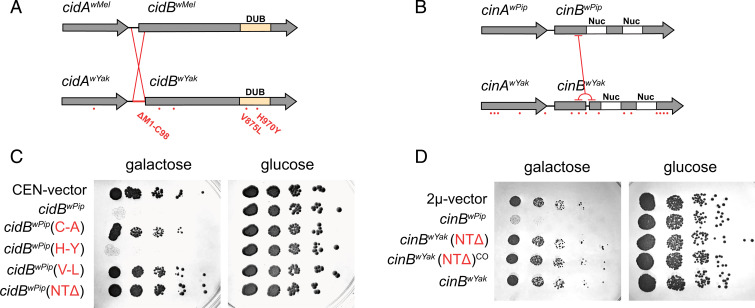
Fig. 1.
Analysis of mutations in yeast model. Analysis of cif permutations in wYak, wMel, and wPip. Amino acid differences between homologs are highlighted with red dots for (A) CidwMel versus CidwYak and (B) CinwPip versus CinwYak. CidBwYak contains two mutations within the DUB domain and a large inversion. CinBwYak contains a tandem duplication yielding a frameshift and premature stop codon but no mutations in the nuclease (Nuc) domains (C). Serial dilutions of yeast-expressing constructs with analog mutations for cid (C) and cin (D). For cid, we express cidBwPip as in previous studies (19), since it produces a strong CI-like phenotype. Introducing (V-L) and (NTΔ) individually reduces yeast toxicity relative to wild-type control, cidBwPip. cidBwPip(C-A) is a catalytic inactive negative control. CEN-vector is an empty pRS416gal1 negative control. For cin, (NTΔ) beginning after the tandem duplication and the wild-type sequence containing the tandem duplication reduces yeast death relative to wild-type control, cinBwPip. (NTΔ) and (NTΔ)CO contain native sequence or codon optimized sequence, respectively. 2μ-vector is an empty pYes2 negative control.