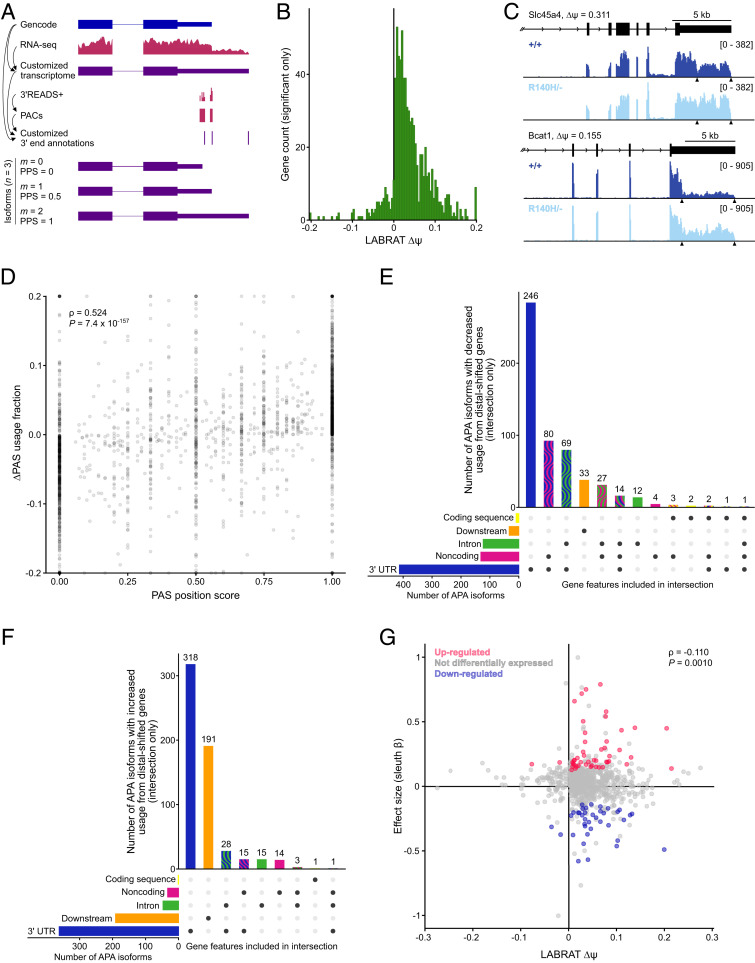
Fig. 4.
PAS selection and gene expression are perturbed in P14 Clp1R140H/− spinal cord. (A, Upper) Workflow for annotation of PACs. A customized reference transcriptome was extrapolated by comparing reference (blue) and experimental (pink) data. (Lower) APA isoforms inferred from the customized reference data. For each gene, APA isoforms are assigned sequential values m corresponding to their rank relative to other APA isoforms, with the APA isoform derived from the use of the most proximal PAS set at m = 0 and the APA isoform from the most distal PAS set at m = n − 1, where n is the total number of APA isoforms for the gene. The PAS position score (PPS) is defined as m ÷ (n − 1). (B) Histogram of genes with significant alternative PAS usage in Clp1R140H/− spinal cord. LABRAT ∆ψ = Clp1R140H/− ψ − Clp1+/+ ψ. ∆ψ values were clipped at ±0.2. (C) Read counts from representative RNA-seq libraries showing the 3′ ends of example genes with significant distal shifts in PAS usage in Clp1R140H/− spinal cord. Reference sequences (mm10) are shown in black and PASs are indicated by arrowheads. (D) Relationship of PAS usage to PAS position for all APA isoforms from genes in which ∆ψ ≠ 0 and P-adjusted ≤ 0.05. PAS usage fraction is the proportion of reads attributed to a given APA isoform relative to the total number of reads associated with the gene. ∆PAS usage fraction = Clp1R140H/− PAS usage fraction − Clp1+/+ PAS usage fraction. Correlation was assessed by Spearman’s rank correlation. Values for ∆PAS usage fraction were clipped at ±0.2. (E and F) UpSet plots showing the numbers of gene features that overlap with the PASs of differentially utilized APA isoforms (∆PAS usage fraction ≠ 0 and P-adjusted ≤ 0.05) from genes with differential PAS usage (LABRAT ∆ψ ≠ 0 and P-adjusted ≤ 0.05). APA isoforms that were derived from distal-shifted genes and show decreased usage in the mutant (E) and APA isoforms that were derived from distal-shifted genes and show increased usage in the mutant (F). (G) Relationship of differential expression to PAS usage for genes with altered PAS usage in Clp1R140H/− spinal cord. Genes in which ∆ψ ≠ 0 and P-adjusted ≤ 0.05 are shown. sleuth β is an approximation of the natural log of the fold-change of expression in Clp1R140H/− spinal cord relative to wild-type spinal cord. Values were clipped at x = ±0.275 and y = ±1. Correlation was assessed by Spearman’s rank correlation. kb, kilobases.