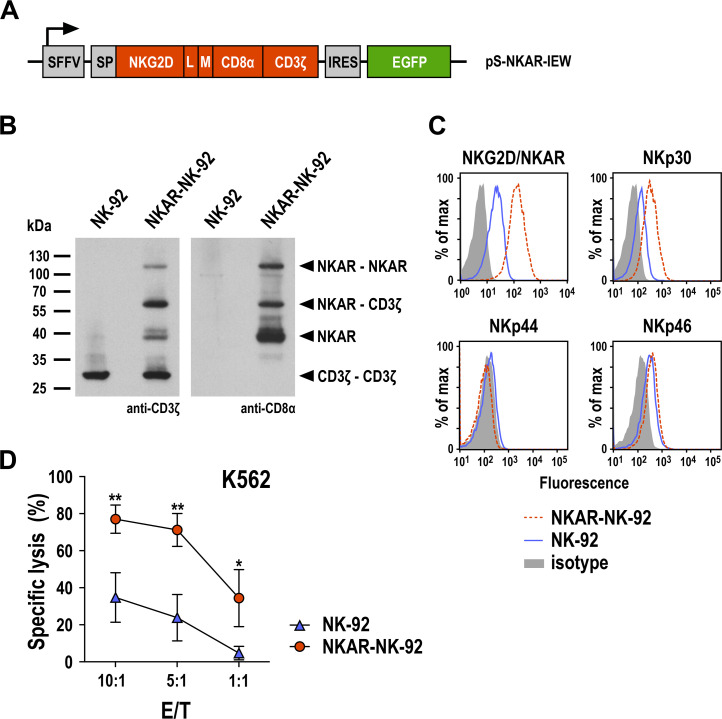
Figure 5.
Generation of NKAR-NK-92 cells. (A) Lentiviral transfer plasmid encoding the NKG2D-based chimeric antigen receptor NKAR under the control of the SFFV promoter. The receptor consists of an immunoglobulin heavy-chain SP, the extracellular domain of NKG2D (amino acid residues 82–216), a flexible (G4s)2 L, an M tag, a CD8α hinge region (CD8α), and transmembrane and intracellular domains of CD3ζ. The NKAR sequence is followed by an IRES and EGFP cDNA. (B) Expression of NKAR by sorted NKAR-NK-92 cells was analyzed by SDS-PAGE of whole cell lysate under non-reducing conditions and immunoblotting with CD3ζ-specific (left) and CD8α-specific antibodies (right), followed by HRP-conjugated secondary antibodies and chemiluminescent detection. Lysate of parental NK-92 cells was included as control. The positions of NKAR homodimers and monomers, CD3ζ homodimers, and NKAR-CD3ζ heterodimers are indicated by arrowheads. (C) Expression of the activating NK-cell receptors NKG2D and NKAR, NKp30, NKp44, and NKp46 in sorted NKAR-NK-92 (dashed red lines) and unmodified parental NK-92 cells (solid blue lines) was analyzed by flow cytometry using receptor-specific antibodies. NK-92 cells stained with irrelevant antibodies of the same isotype served as controls (filled areas). (D) Cytotoxicity of NKAR-NK-92 (red circles) and parental NK-92 cells (blue triangles) against K562 erythroleukemia cells was investigated in flow cytometry-based cytotoxicity assays after coincubation at different E/Ts for 3 hours. Mean values±SD are shown; n=3 technical replicates from a representative experiment. Data were analyzed by two-tailed unpaired Student’s t-test. *P<0.05, **P<0.01. EGFP, enhanced green fluorescent protein; E/T, effector to target ratio; IRES, internal ribosome entry site; L, linker; M, Myc; NK, natural killer; NKG2D, natural killer group 2D; SFFV, spleen focus-forming virus; SP, signal peptide.