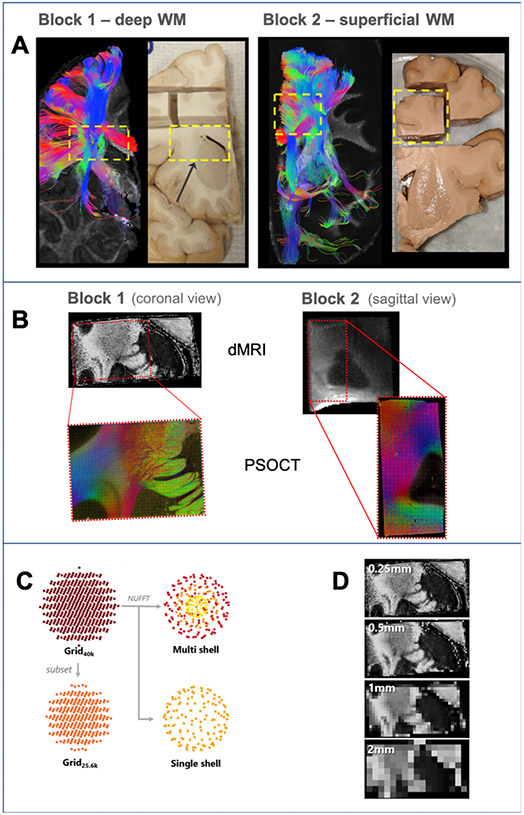
Fig. 1. Overview of experiments.
A: For each brain, we used deterministic DTI tractography in a whole-hemisphere scan to identify a block that we extracted for further scanning and analysis (yellow dashed box). B: dMRI data of the blocks were acquired at 9.4 T (FA maps, top) and a region of each block (red box) was excised for imaging with PSOCT (RGB fiber orientation maps, bottom). C: Different dMRI sampling schemes investigated. The data were acquired on a grid in q-space (Grid40k). An additional dataset was created from a low-q subset of the original data (Grid25.6k). The nonuniform fast Fourier transform (NUFFT) was used to generate multi- and single-shell datasets. D: Spatial resolutions included in dMRI analysis. The acquired 0.25 mm resolution data were retrospectively downsampled to generate lower spatial resolution data.