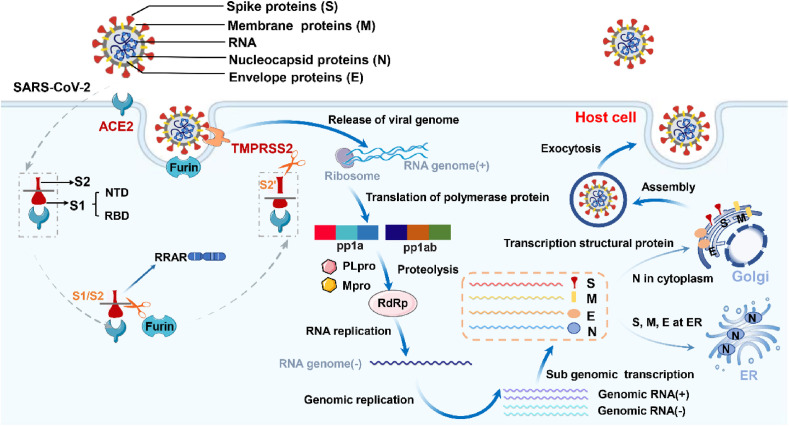
Fig. 1.
The life cycle of SARS-CoV-2. When SARS-CoV-2 S protein binds ACE2 of the host cell with a high affinity, the virus attaches to the cell surface and destroys the stability of the trimer before S protein fusion. S protein cleavage usually occurs sequentially, with furin enzyme cleavage at the S1/S2 site first, followed by TMPRSS2 cleavage at the S2’ site. Under protease cleavage, the S1 (RBD)–ACE2 complex was separated from the S2 subunit, and SARS-CoV-2 enters cells through endocytosis. SARS-CoV-2 releases the single chain positive RNA, then translates into pp1a and pp1ab with the help of ribosomes of host cells, and produces hydrolytic enzymes that can precisely cut polyprotein by means of self-shearing, namely main protease (Mpro) and papain-like protease (Plpro). Under the hydrolysis of Mpro and Plpro, RNA dependent RNA polymerase (RdRp) was formed. RdRp synthesized RNA genome (−) and then synthesized virus genome through genome replication. N protein binds to genomic RNA and replicates, transcribes and synthesizes in the cytoplasm. S, M and E proteins integrate into the membrane of the endoplasmic reticulum (ER). Virus are transported to the host cell membrane and released by exocytosis, and then infect other tissues and cells.