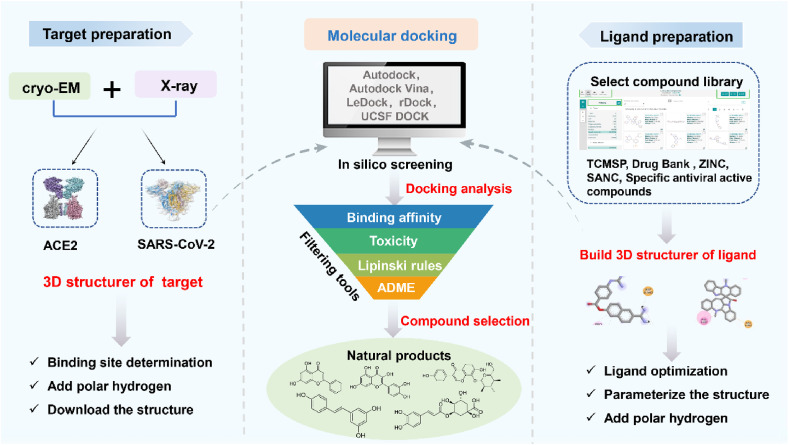
Fig. 2.
Molecular docking process. Cryo-EM and X-ray reveal the structure of SARS-CoV-2 and ACE2. Downloading the structure and manually removing all binding ligands, ions and solvent molecules from the protein database for parameterization, and constructing 3D structure of targets. The ligands are mainly obtained from TCMSP, Drug Bank, ZINC, SANC and other databases. Then, Autodock tools is used to parameterize the structure to add complete hydrogen to the ligands. Finally, the ligands were subjected to molecular docking with SARS-CoV-2 and ACE2. The toxicity and pharmacokinetics of the compounds were analyzed. The ADME (Absorption, Distribution, Metabolism, and Excretion) properties of the compounds were evaluated by means of mathematical algorithm, and the bioavailability of compounds could be predicted by the most classic Lipinski rule to further obtain the potential drug-forming compounds with activity.