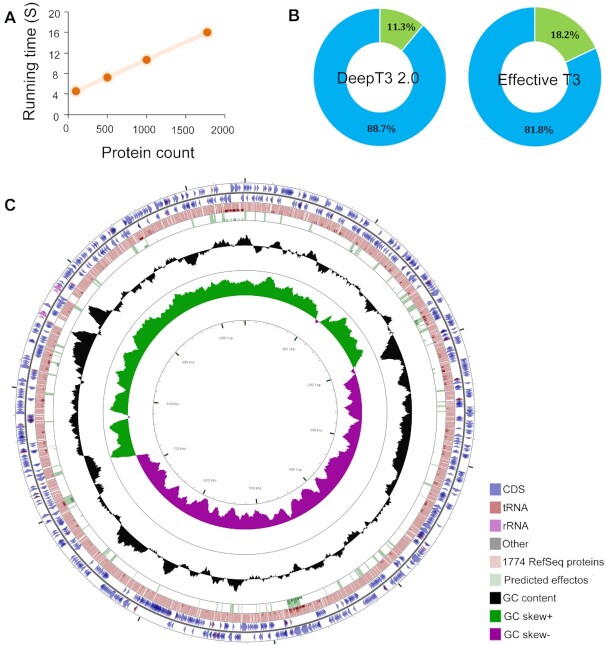
Figure 6.
The application of DeepT3 2.0 to whole-genome prediction in the Chlamydia trachomatis genome. (A) Run times of DeepT3 2.0 for inputs of varying sizes. (B) Detailed proportions of predicted effector for two T3SE prediction methods. (C) Visualizing features, ORFs, start and stop codons of Chlamydia trachomatis genome and comparing all RefSeq proteins and DeepT3 2.0 predicted effectors. The proteins are mapped according to their corresponding positions on the circular bacterial genome.