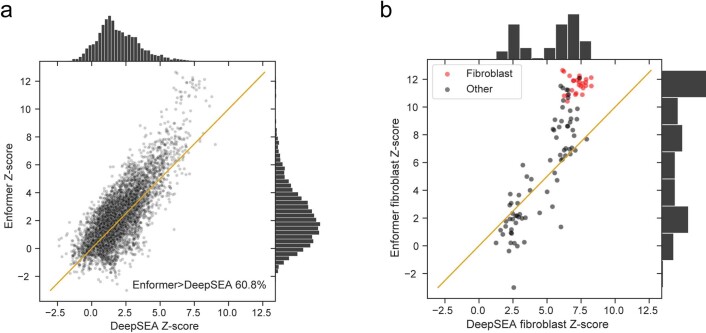
Extended Data Fig. 9. Enformer achieves greater and more specific SLDP concordance to GTEx than DeepSEA.
To compare Enformer and DeepSEA Beluga (convolutional neural network used in ExPecto) variant effect predictions, we manually matched DNase datasets that both models were trained on, finding 100 confident matches. We computed genome-wide statistical concordance between variant effect predictions for these DNase datasets and GTEx eQTL summary statistics using SLDP across all variants in the 1000 genomes dataset. a) We scatter plotted all DNase sample and GTEx tissue z-scores for the DeepSEA and Enformer predictions, observing that the Enformer scores are greater for 60.8%. Each point corresponds to (DNase sample, GTEx tissue) pair. Only some (DNase sample, GTEx tissue) pairs are biologically well-matched, while the majority are. b) Since many DNase samples profile fibroblasts we specifically plotted all DNase sample z-scores for the GTEx fibroblast summary statistics. We colored the DNase fibroblast samples in red, revealing that they are the highest scoring and most improved in the Enformer model relative to DeepSEA. This suggests that Enformer variant effect predictions are more tissue specific, since one would expect to obtain the highest Z-score for these matched samples shown in red.