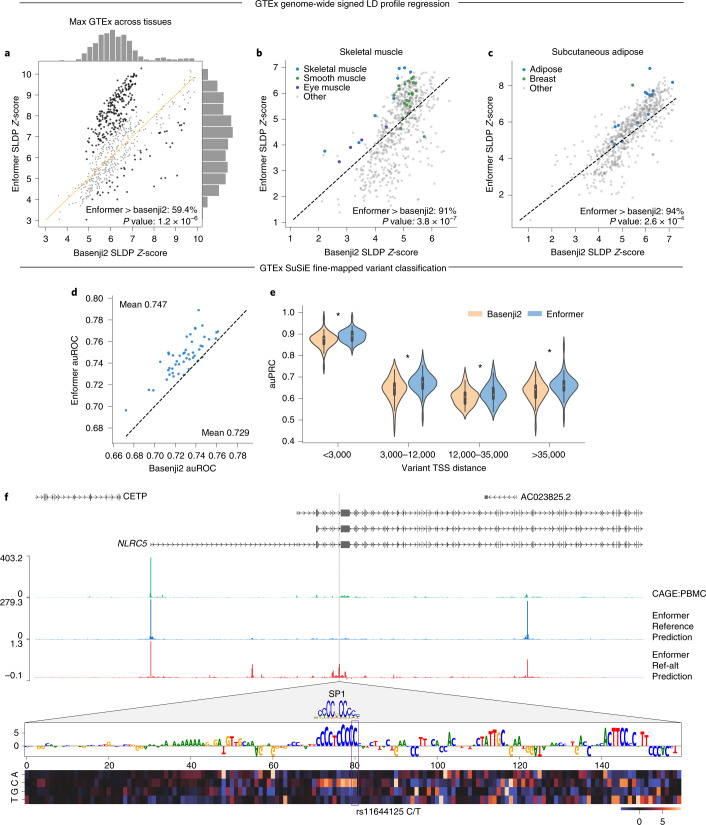
Fig. 3. Enformer improves variant effect prediction on eQTL data as measured by SLDP regression and fine-mapped variant classification.
a, We computed genome-wide statistical concordance between variant effect predictions for individual CAGE datasets and GTEx eQTL summary statistics using SLDP21 across all variants in the 1000 Genomes dataset. Taking the GTEx tissue with max Z-score for each sample, Enformer predictions achieved greater Z-scores for 59.4% of samples, and 228 are greater by more than one s.d. (versus 46 for Basenji2). Each point represents one of the 638 CAGE samples. We used one-sided Binomial tests to compute the P values in the top row panels. b,c, Studying SLDP in skeletal muscle (b) and subcutaneous adipose (c) GTEx tissues indicated that biologically relevant CAGE datasets (shown in blue) improve between Basenji2 and Enformer. d, We trained random forest classifiers to discriminate between fine-mapped GTEx eQTLs and matched negative variants in each of 48 tissues (Methods). Features derived from Enformer enabled more accurate classifiers than Basenji2 features for 47 of 48 tissues. e, We computed auPRC for variants in four roughly equally sized TSS distance bins. Violin plots represent measures for the n = 48 tissues (white dots represent the median, thick bars the interquartile range, and thin bars the entire data range). Enformer improved accuracy at all distances (one-sided paired Wilcoxon P < 1 × 10–4). f, Enformer prediction for rs11644125 improved relative to Basenji2 (data not shown) by better capturing its influence on an NLRC5 TSS ~35 kb upstream. rs11644125 is associated with monocyte and lymphocyte counts in the UK BioBank and fine-mapped to >0.99 causal probability24. In silico mutagenesis of the region surrounding rs11644125 revealed an affected SP1 transcription factor motif39.