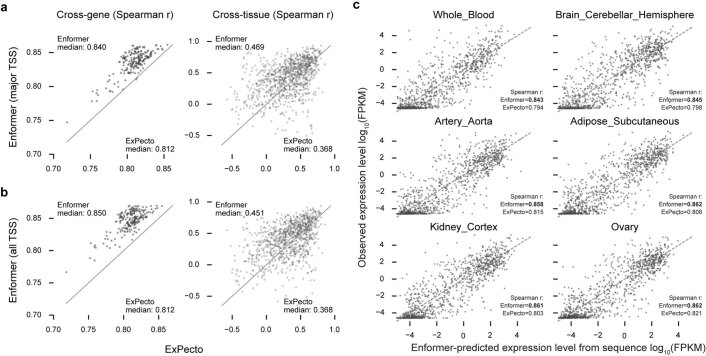
Extended Data Fig. 4. Enformer predicts mRNA-seq more accurately than ExPecto.
a) Test set predictive performance comparison of a linear model trained on top of Enformer CAGE predictions from the major TSS (y-axis) and ExPecto (x-axis) computed either across genes (first column) or across tissues (second column). Gene expression matrix was normalized across genes to have zero mean and unit variance for each tissue. Enformer was re-trained only on the human genome using the same training chromosomes for this comparison (Methods). b) Same as a), but using Enformer predictions averaged across all TSS of the gene. c) Observed versus Enformer-predicted gene expression values for all 990 test genes in 6 RNA-seq samples.