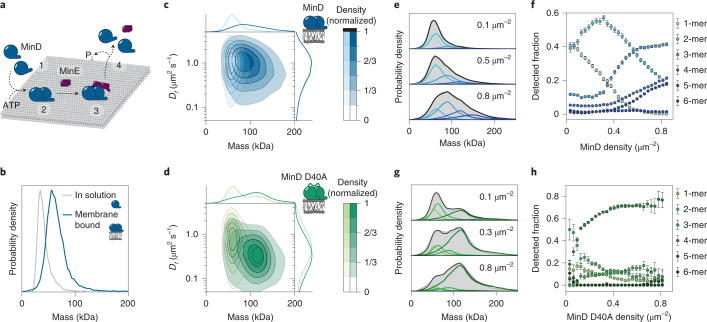
Fig. 2. Lateral MinD–MinD interactions lead to self-assembly into large homo-oligomers.
a, Schematic of the canonical membrane binding–unbinding cycle of MinDE. Upon ATP complexation, MinD dimerizes (1) and attaches to the membrane interface (2). In the event of MinE binding (3), MinE stimulates the intrinsic ability of MinD to hydrolyze ATP, which upon inorganic phosphate (Pi) release leads to the dissociation of MinD from the membrane in its monomeric form (4). b, MinD mass distribution in solution (gray line) (n = 16,101 particles) and on attachment to the SLB (blue line) (n = 13,917 trajectories). For solution experiments, 175 nM MinD with 0.5 µM ATP were measured in the conventional mass photometry landing assay. The membrane mass distribution of MinD was determined using MSPT at a particle density of 0.03 µm−2. c,d, Two-dimensional KDE of membrane-attached MinD (c) and MinD D40A (d) at particle densities of 0.1 µm−2 (light blue) (n = 117,086 trajectories) and 0.8 µm−2 (dark blue) (n = 152,685 trajectories) and 0.1 µm−2 (light green) (n = 7,831 trajectories) and 0.8 µm−2 (dark green) (n = 3,150 trajectories), respectively. Marginal probability distributions of both molecular mass (top) and diffusion coefficient (right) are presented. e,g, Representative mass distributions (gray) of MinD (e) and MinD D40A (g) and estimation (black line, colored lines highlight underlying components) of its six components (MinD monomer–hexamer, light blue–dark blue; MinD D40A monomer–hexamer, light green–dark green) for three different particle densities (0.1 µm−2, 0.3/0.5 µm−2 and 0.8 µm−2). f,h, Relative oligomer abundance as a function of particle density: MinD (f) and MinD D40A (h). Error bars are the standard deviation of fitting results from three data subsets. The oligomer analysis is based on a total of n = 1,102,940 trajectories for MinD and n = 194,545 for MinD D40A.