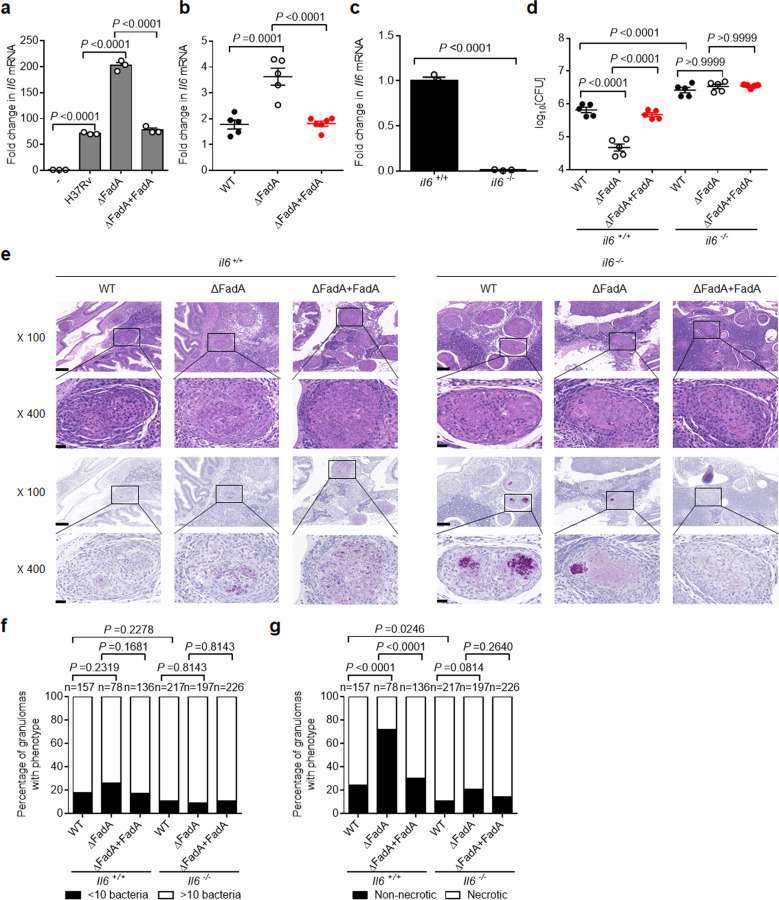
Fig. 3. FadA inhibits host immune responses.
a qPCR analysis of Il6 mRNA from peritoneal macrophages infected with H37Rv, H37RvΔFadA, or H37Rv(ΔFadA + FadA) strains for 4 h (MOI = 1) (mean ± SEM). b qPCR analysis of Il6 mRNA from adult zebrafish intraperitoneally infected with roughly 200 CFU per fish of WT, ΔFadA, or ΔFadA + FadA M. marinum strains for 14 days (mean ± SEM of n = 5). c qPCR analysis of Il6 mRNA from il6+/+ and il6−/− zebrafish (mean ± SEM). The Cas9/gRNA system was employed to generate IL-6 knockout zebrafish, which were constructed at the China Zebrafish Resource Center (CZRC) as described previously. d–g il6+/+ and il6−/− mutant adult zebrafish were intraperitoneally infected with approximately 200 CFU of WT, ΔFadA, or ΔFadA + FadA M. marinum strains for 14 days. Bacterial titers (d; mean ± SEM of n = 5 fish infected for 14 days), histopathology (e; representative of one experiment with at least three independent replicates; scale bar, 100 μm (top) and 20 μm (bottom)), and comparison of granulomas between WT, ΔFadA, or ΔFadA + FadA M. marinum-infected il6+/+ and il6−/− adult zebrafish scored for M. marinum burden as less than 10 or 10 or more bacteria (f) or percentage of necrotic granulomas in each fish (g) were assessed as described previously. “n” was the total number of granulomas for each strain infected fish. Total number of zebrafish analyzed: five (WT/il6+/+), five (ΔFadA/il6+/+), five (ΔFadA+FadA/il6+/+), five (WT/il6−/−), five (ΔFadA/il6−/−), five (ΔFadA + FadA/il6−/−). Data in a–g represent one experiment with at least three independent replicates. One-way ANOVA with Bonferroni’s multiple comparisons test (a, b, d), two-tailed unpaired Student’s t-test (c) and Fisher’s exact test (f, g) were used for statistical analysis.