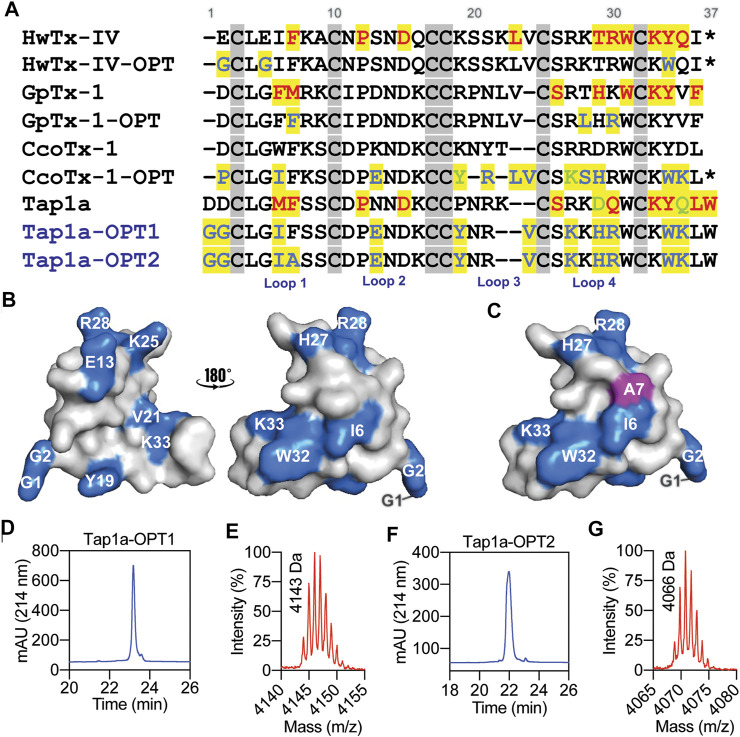
FIGURE 2.
Rational design of Tap1a-OPT1 and Tap1a-OPT2, and peptide production via recombinant expression. (A) Alignment of NaSpTx1 spider peptides which were pharmacologically optimized for NaV channels inhibition (Cardoso and Lewis, 2019), and Tap1a, Tap1a-OPT1 and Tap1a-OPT2. Residues coloured in red are positions where substitutions led to NaV-activity loss reported in previous studies and for Tap1a in this work, and residues coloured in blue and green are substitutions that enhanced potency and selectivity for NaVs, respectively. Residue substitutions in Tap1a-OPT1 and Tap1a-OPT2 are highlighted in blue. Cysteines are in grey boxes. (B, C) Model of the three-dimensional structures of Tap1a-OPT1 (B) and Tap1a-OPT2 (C) highlighting in blue the substitutions introduced in Tap1a-OPT1 and in pink the additional substitution F7A introduced in Tap1a-OPT2. (D–G) Production of the peptides Tap1a-OPT1 and Tap1a-OPT2. RP-HPLC chromatogram and mass spectrometry analysis results of the purified recombinants Tap1a-OPT1 (D, E) and Tap1a-OPT2 (F, G) purified in C18 column using a TFA/ACN gradient. Mass spectrometry analysis of purified peptides displayed the expected masses of 4143 Da for Tap1a-OPT1 (E) and 4066 Da for Tap1a-OPT2 (G).