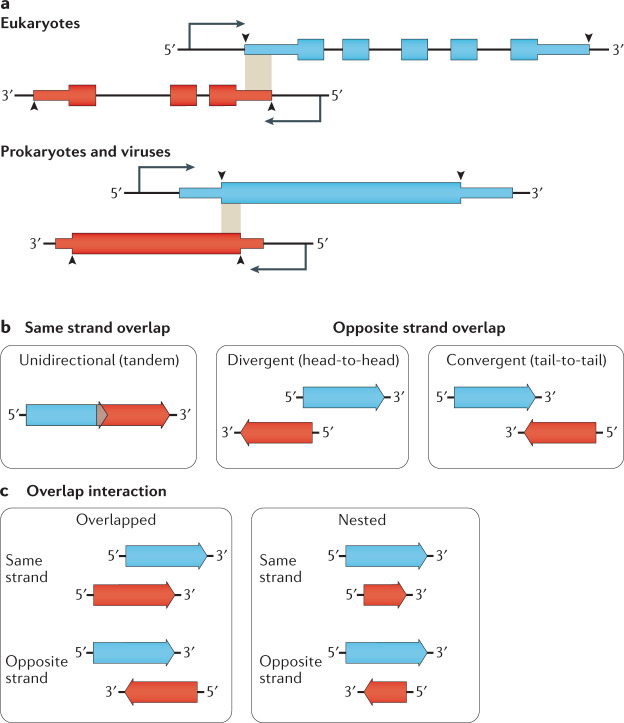
Fig. 1. Overlapping gene definition and topologies.
a | Gene overlap definitions differ between prokaryotes and eukaryotes. (Top) Eukaryote overlaps are most frequently defined as overlaps between the boundaries of the primary transcript, shown here in the shaded region. Often, the overlap is only between the 5′ untranslated region (UTR) or 3′ UTR of both transcripts (5′ UTR overlap shown)10,170. (Bottom) In contrast, prokaryote and virus genes are only considered to overlap if their coding sequences overlap5,27. Thin boxes denote 5′ and 3′ UTRs while thick boxes are coding sequences. Arrowheads indicate the extent of the consensus definition of gene boundaries within studies referenced in this review. b | Genes and open reading frames (ORFs) can be overlapped in one of three topologies. Unidirectional (also called tandem) overlaps occur between genes and ORFs on the same strand. Divergent (also called head-to-head) overlaps occur between genes and ORFs on opposite strands that overlap at their 5′-ends. Convergent (also called tail-to-tail) overlaps occur between genes and ORFs on opposite strands that overlap at the 3′-ends27. c | Gene and ORF interactions can be either overlapped, where only limited portions of each gene or ORF are overlapping, or nested, where the entire sequence of one partner falls within the boundaries of the other.