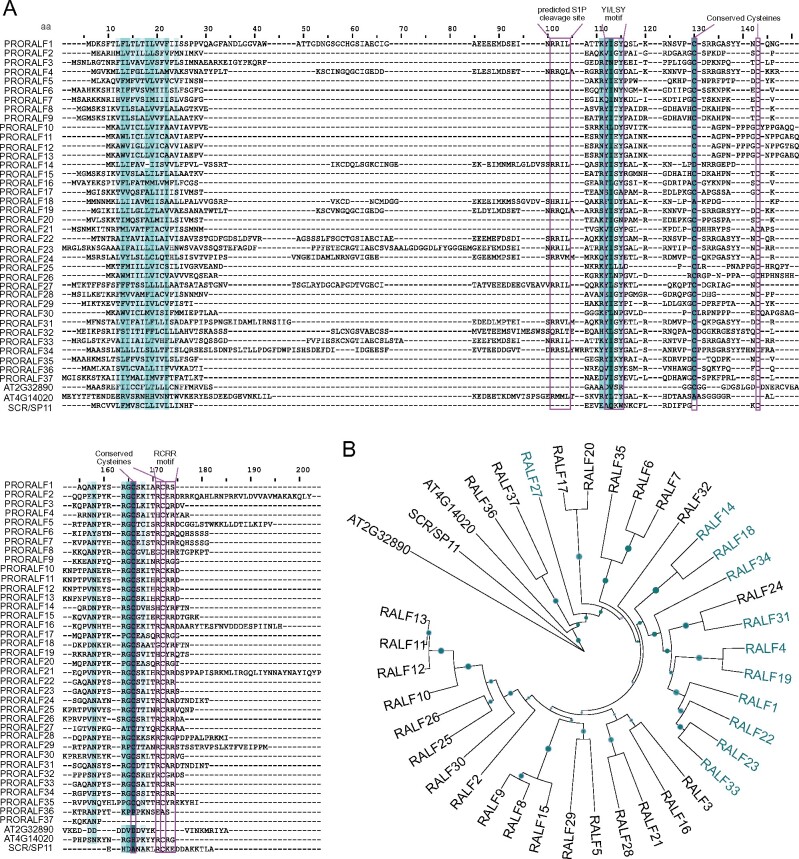
Figure 1.
Re-evaluation of the Arabidopsis RALF family. A, Alignment of AtPRORALFs, AT2G32890, and AT4G14020. Color-code based on sequence conservation: the darker the color, the more conserved the residue. Pink boxes indicate conserved motifs. B, Rooted phylogenetic tree of the AtRALF peptides, AT2G32890 and AT4G14020. UPGMA tree inferred from the MUSCLE alignment displayed in Figure 1, A. S-locus protein 11 or S-locus Cys-rich (SCR/SP11) sequence was used to root the tree. RALFs highlighted in teal indicate those predicted to be cleaved by the protease S1P. Bootstrap values (1,000 repetitions) above 50% are represented by green circles in the corresponding branches. The higher the bootstrap value for a particular branch, the higher the size of the circle.