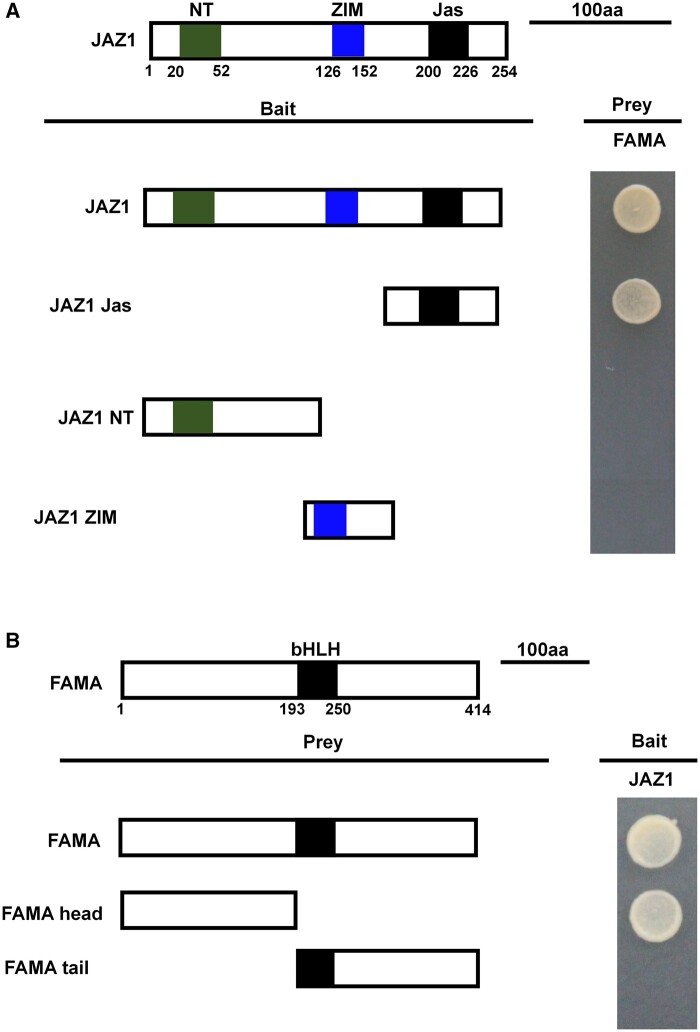
Figure 4.
Mapping of the protein domains involved in the interaction of FAMA with JAZ1, using Y2H assays. A, Based on the schematic protein structure of JAZ1, full-length JAZ1, or its derivatives (pGBKT7-JAZ1 or pGBKT7-JAZ1 derivatives) were tested for interactions with FAMA (pGADT7-FAMA). Yeast cells cotransformed with pGBKT7-JAZ1 or pGBKT7-JAZ1 derivatives (baits) and pGADT7-FAMA (prey) were grown on yeast synthetic dropout lacking Leu and Trp (SD/-2) as transformation control, or on selective media lacking Ade, His, Leu, and Trp (SD/-4) to test protein interactions. The different truncations of JAZ1 are represented. B, Based on the schematic protein structure of FAMA, full-length FAMA, or its derivatives (pGADT7-FAMA or pGADT7-FAMA derivatives) were tested for interactions with JAZ1 (pGBKT7-JAZ1). Yeast cells cotransformed with pGADT7-FAMA or pGADT7-FAMA derivatives (prey) and pGBKT7-JAZ1 (bait) were grown on yeast synthetic dropout lacking Leu and Trp (SD/-2) as transformation control, or on selective media lacking Ade, His, Leu, and Trp (SD/-4) to test protein interactions. The different domains of FAMA are represented.