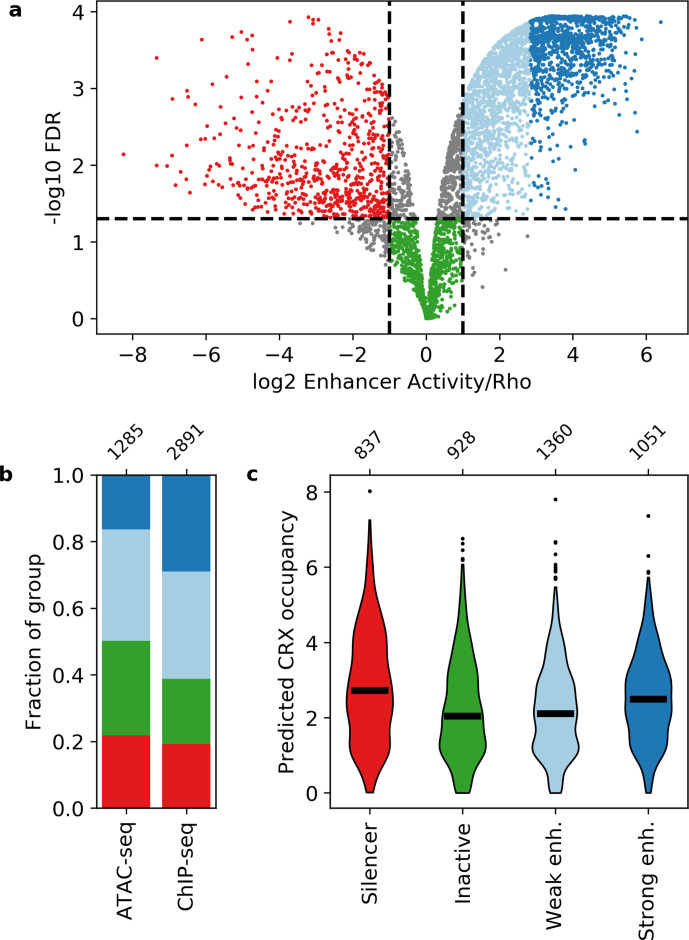
Figure 1. Activity of putative cis-regulatory sequences with cone-rod homeobox (CRX) motifs.
(a) Volcano plot of activity scores relative to the Rho promoter alone. Sequences are grouped as strong enhancers (dark blue), weak enhancers (light blue), inactive (green), silencers (red), or ambiguous (gray). Horizontal line, false discovery rate (FDR) q = 0.05. Vertical lines, twofold above and below Rho. (b) Fraction of ChIP-seq and ATAC-seq peaks that belong to each activity group. (c) Predicted CRX occupancy of each activity group. Horizontal lines, medians; enh., enhancer. Numbers at top of (b and c) indicate n for groups.