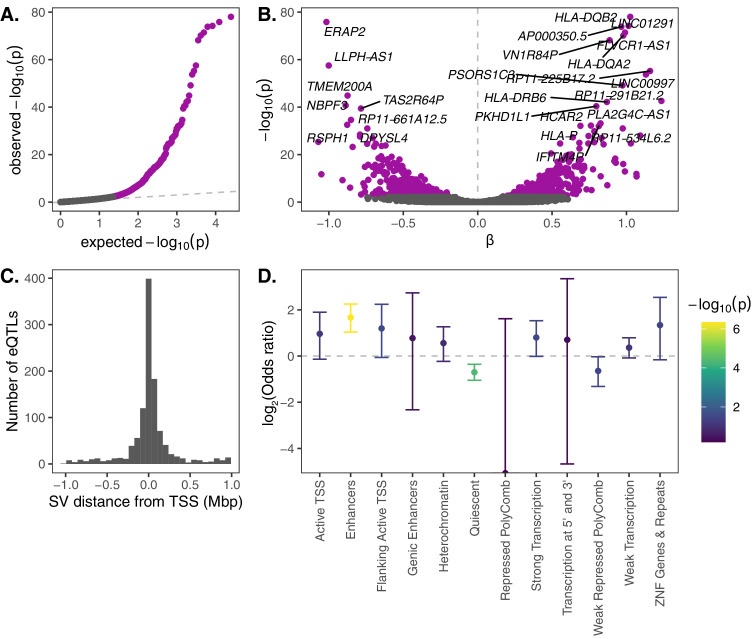
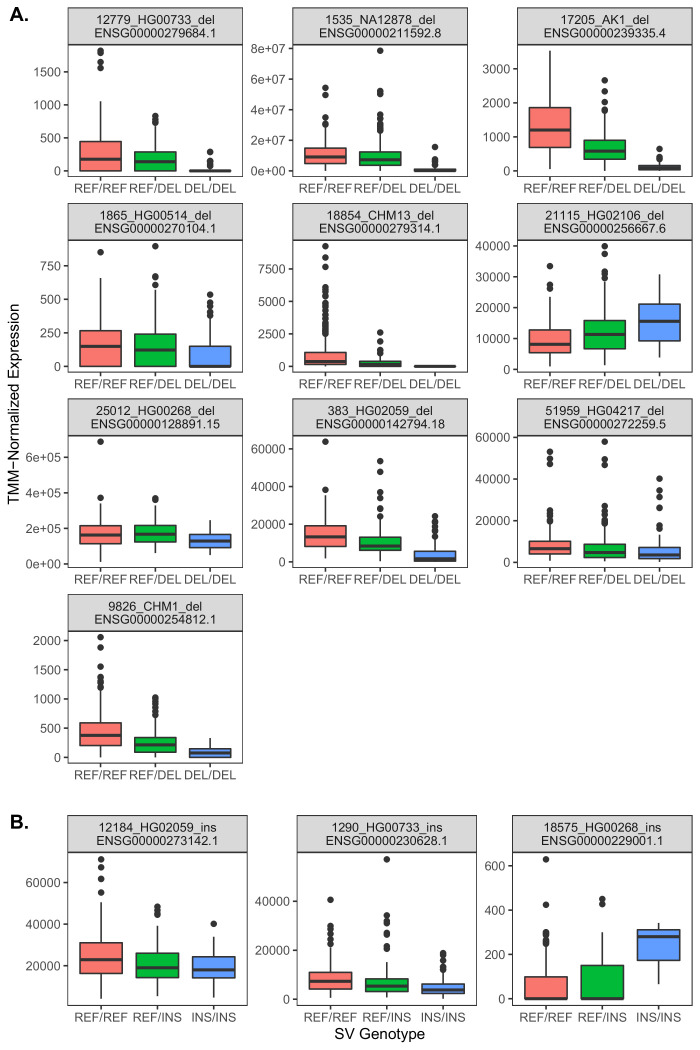
Figure 2. eQTL mapping of SVs.
We used RNA-seq data from the Geuvadis Consortium (Lappalainen et al., 2013), obtained from LCLs derived from individuals from four European and one African population of the 1KGP dataset, to test for associations between SV genotypes and gene expression. SV-eQTL pairs that were significant at a 10% FDR are depicted in purple. (A) Q–Q plot of permutation p-values for all SV-gene pairs tested. (B) Volcano plot of eQTLs and the estimated effect of the alternative allele on expression (β). (C) Distribution of the distance of significant SV eQTLs from the transcription start site (TSS) of their associated genes. (D) Enrichment or depletion of expression associations for SVs that overlap various ChromHMM chromatin state annotations from the Roadmap Epigenomics Project (GM12878 Lymphoblastoid Cells). Chromatin states with ≤0.1% genome-wide representation were omitted for visual clarity.