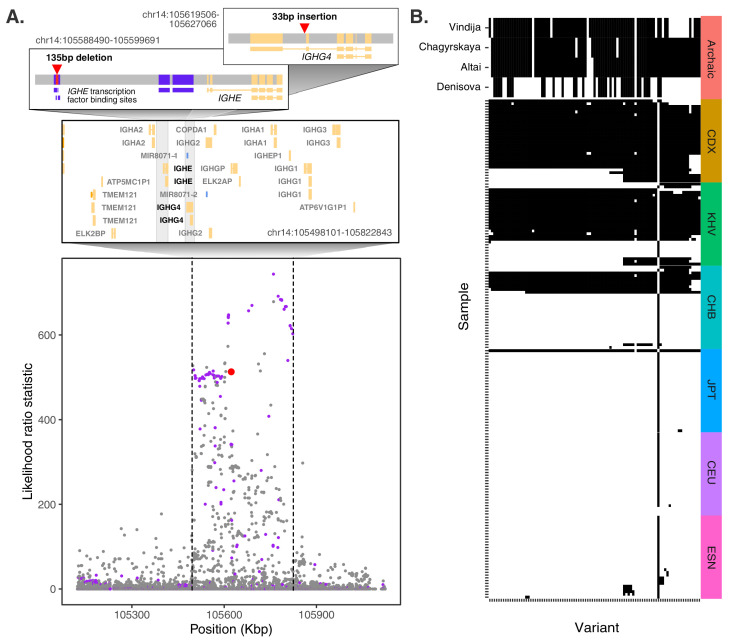
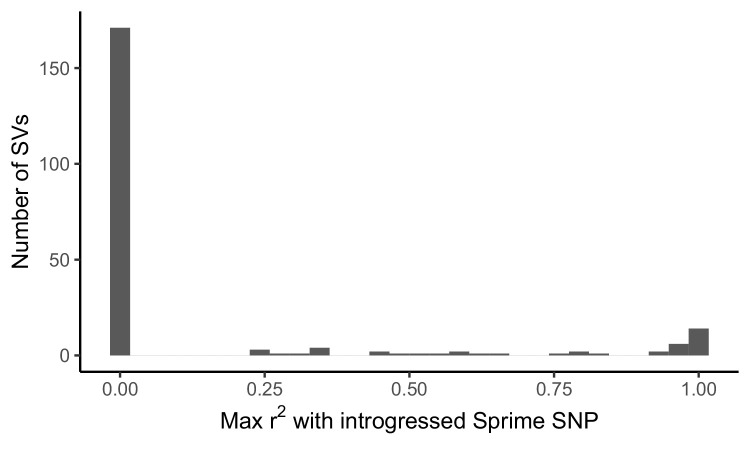
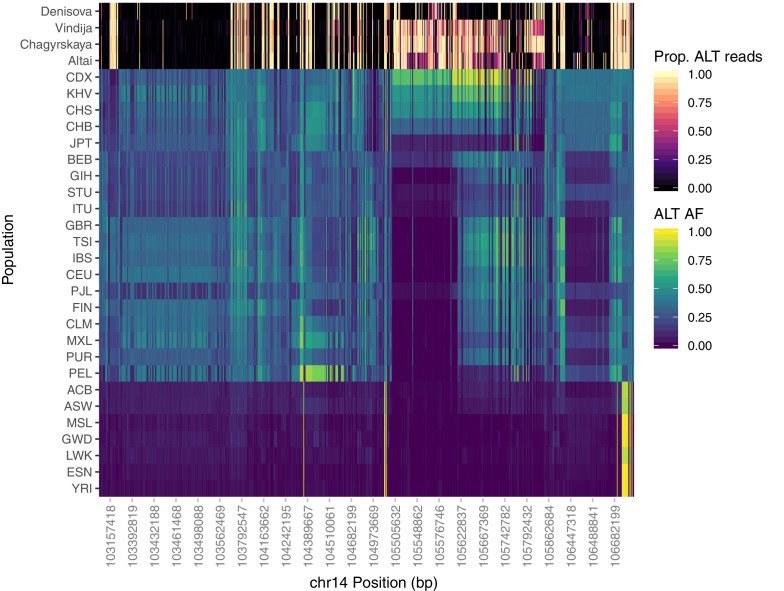
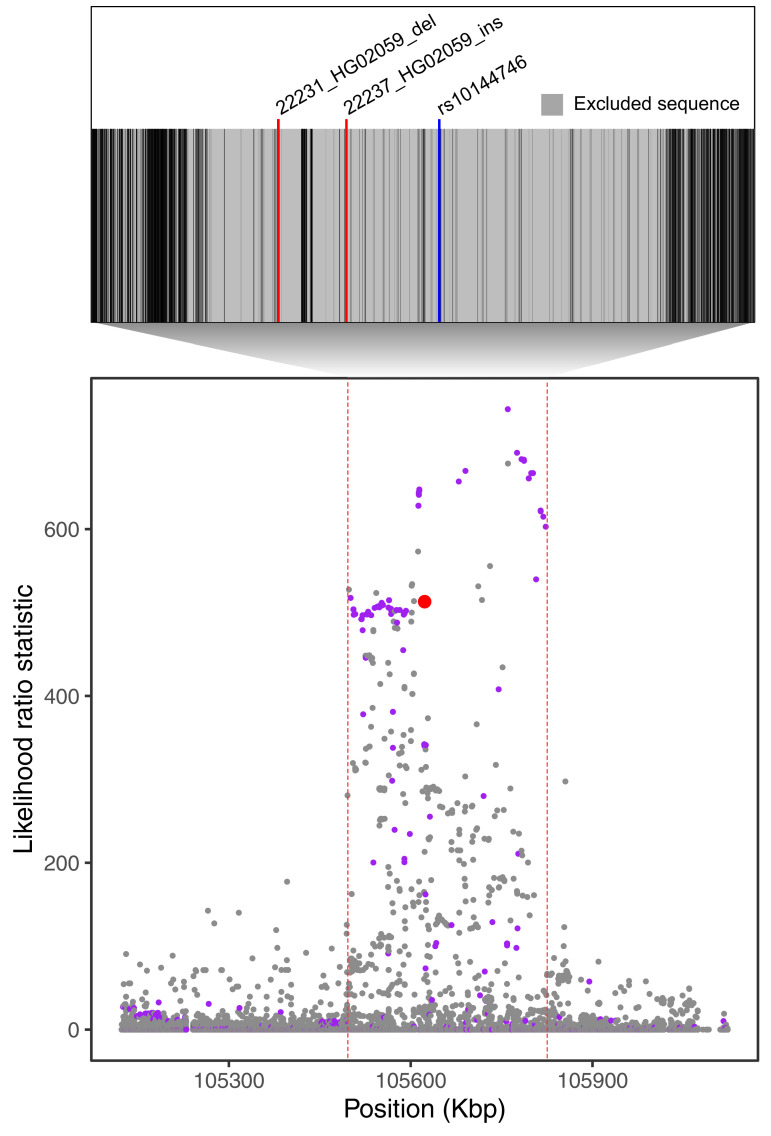
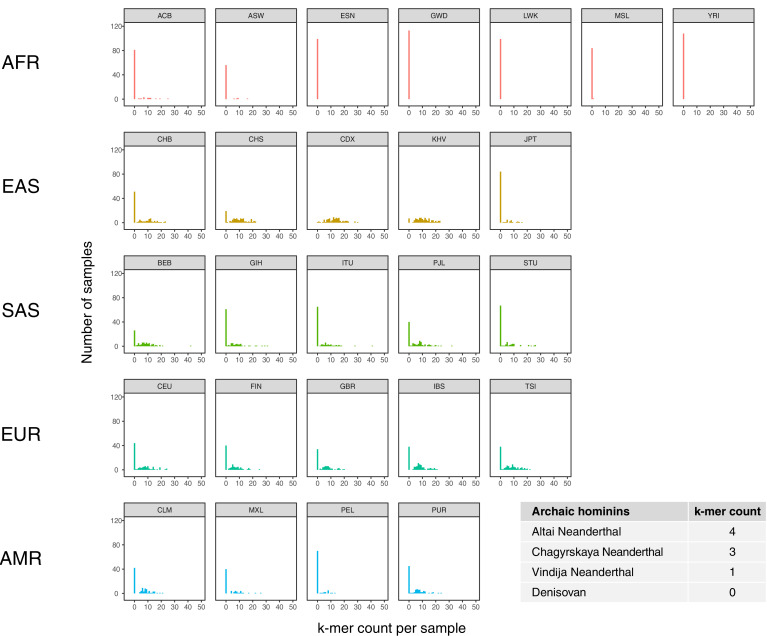
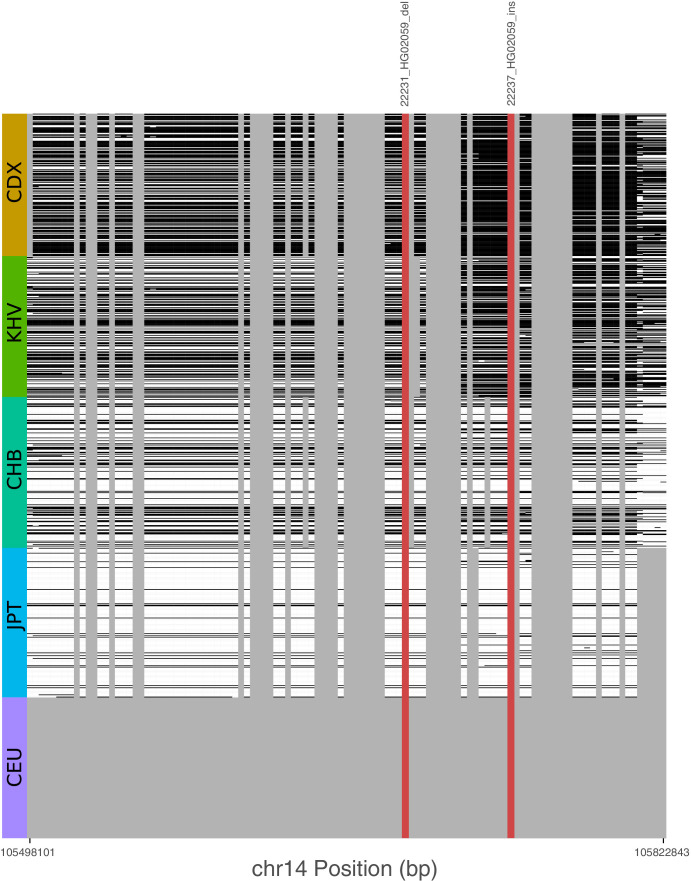
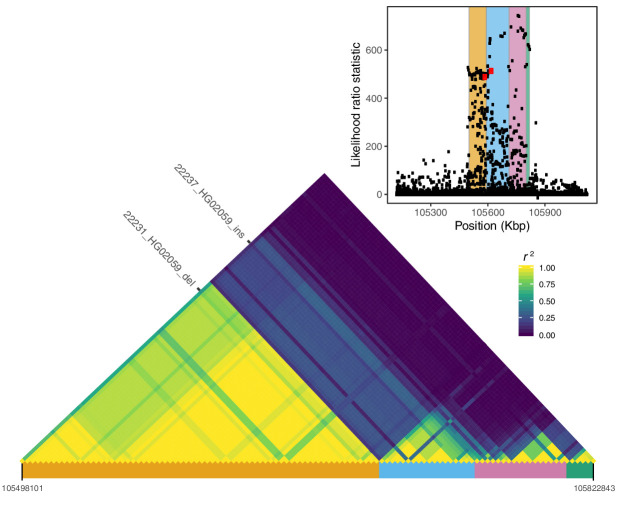
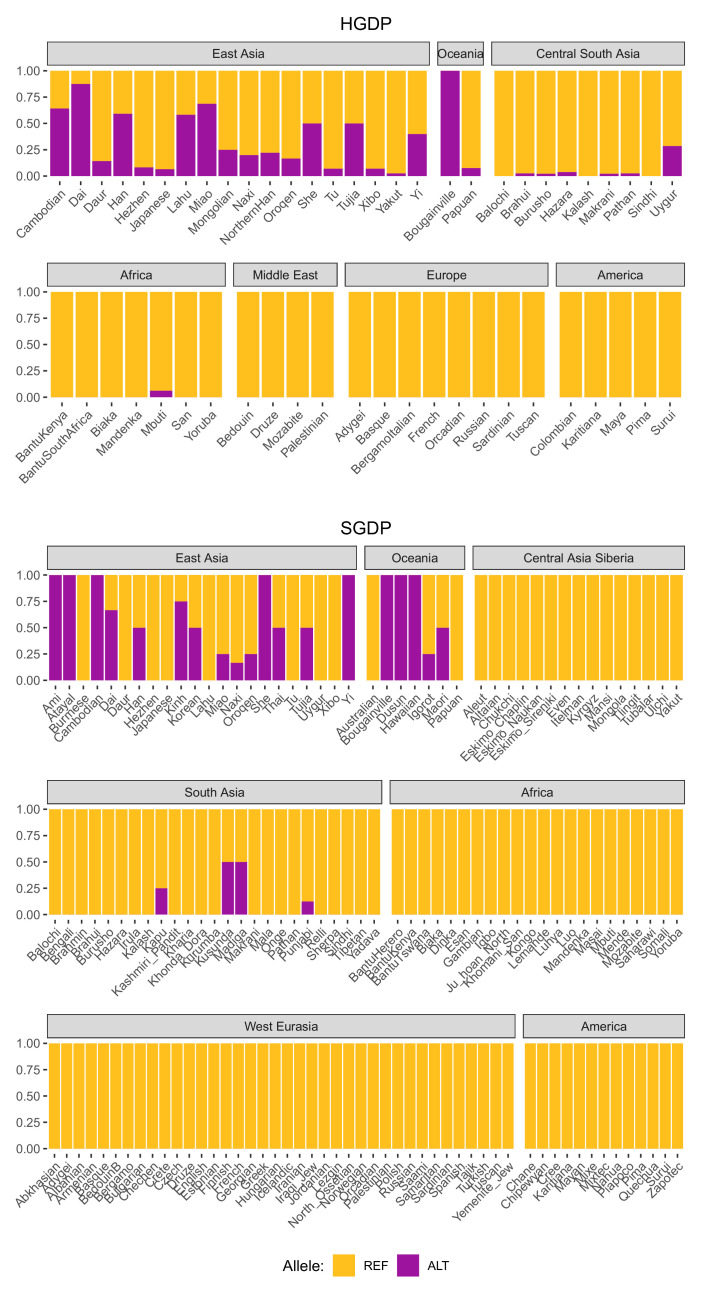
Figure 4. Evidence for Neanderthal introgression of the adaptive IGH haplotype.
(A) Local analysis of likelihood ratio statistics (LRS) in the region near the 33 bp insertion (red point) reveals a 325 kb haplotype encompassing 94 SNPs with strong allele frequency differentiation within ancestry component 2. Points where the alternative allele matches an allele observed in the Chagryskaya Neanderthal genome but at a frequency of 1% or less in African populations are highlighted in purple. (B) Individual haplotypes defined by the highly differentiated SNPs (LRS > 450). Four archaic hominin genomes are plotted at the top, while 30 randomly sampled haplotypes from each of 6 populations from 1KGP are plotted below. Archaic hominins samples are colored according to whether they possess more than one aligned read supporting the alternative allele at a given site. ESN refers to the Esan in Nigeria population of 1KGP. Other 1KGP population codes are provided in the main text.