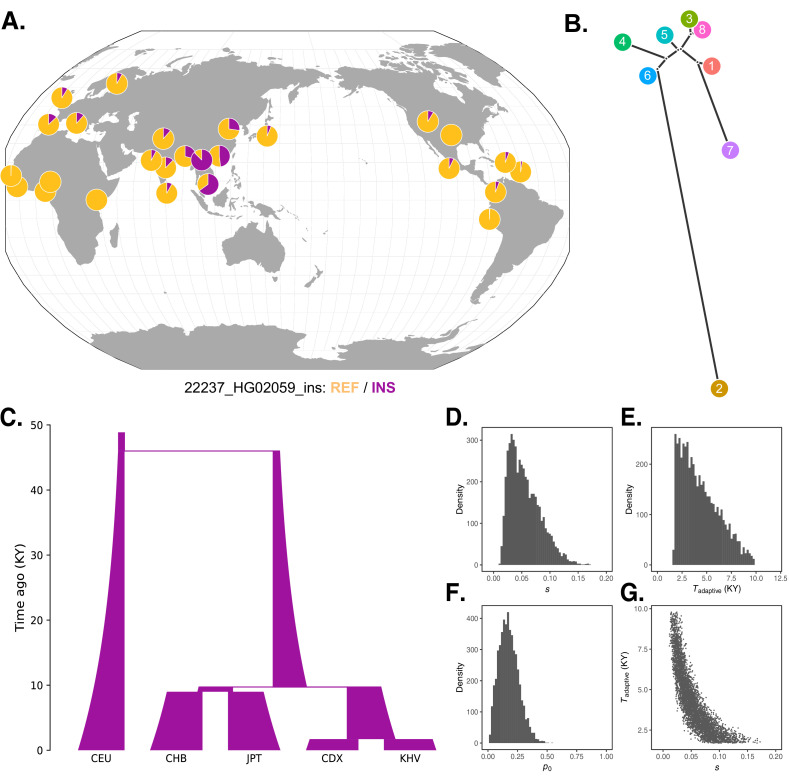
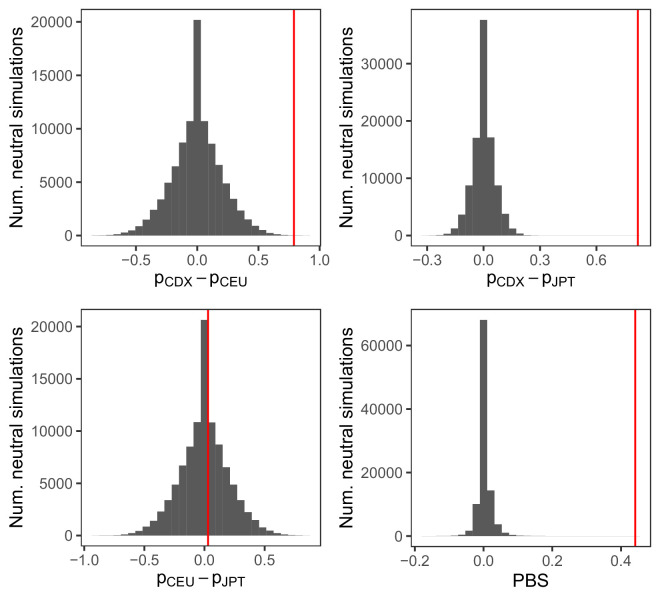
Figure 5. Local adaptation at the IGH locus.
(A) Population-specific frequencies of the putative Neanderthal-introgressed insertion allele in each of the 1KGP populations, in the style of the Geography of Genetic Variants browser (Marcus and Novembre, 2017). (B) Tree representation of the best-fit selection hypothesis for Neanderthal-introgressed haplotype tagged by the IGHG4 insertion polymorphism, as computed by Ohana. (C) Five-population demographic model used for simulation and parameter inference via approximate Bayesian computation (ABC). Population sizes and split times are further described in Materials and methods. (D) Posterior distribution of the selection coefficient (s). (E) Posterior distribution of the timing of the onset of selection (Tadaptive). (F) Posterior distribution of the initial allele frequency at the beginning of the simulation (p0). (G) Negative relationship between s and Tadaptive for simulations retained by ABC.