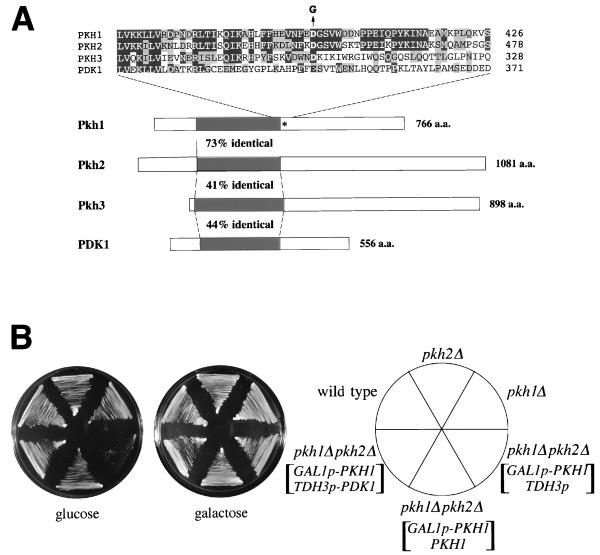
FIG. 2.
Identification of yeast PDK1 homologs. (A) Schematic diagrams of the structures of PDK1, Pkh1, Pkh2, and Pkh3. Kinase domains are indicated by black boxes. The mutated residue in pkh1D398G is indicated by an asterisk. The amino acid (a.a.) residues in the region of the mutation site are aligned above the diagram. The Asp-to-Gly change in the pkh1D398G mutation is indicated as a G above the Pkh1 sequence. Amino acids which are identical or conserved are indicated by black or gray boxes, respectively. (B) Effect of the pkh1Δ and pkh2Δ mutations on cell growth. Yeast strains carrying the indicated plasmids were streaked onto YPGlu (glucose) medium and YPGal (galactose) medium and incubated at 30°C. Yeast strains were 15Dau (wild type), INA25-3B (pkh1Δ::URA3), INA28-1B (pkh2Δ::URA3), and INA38-6A (pkh1Δ::URA3 pkh2Δ::URA3). Plasmids were YCpG22-PKH1 (GAL1p-PKH1), pKT10 (TDH3p), YCplac22-PKH1 (PKH1), and pKT10-PDK1 (TDH3p-PDK1).