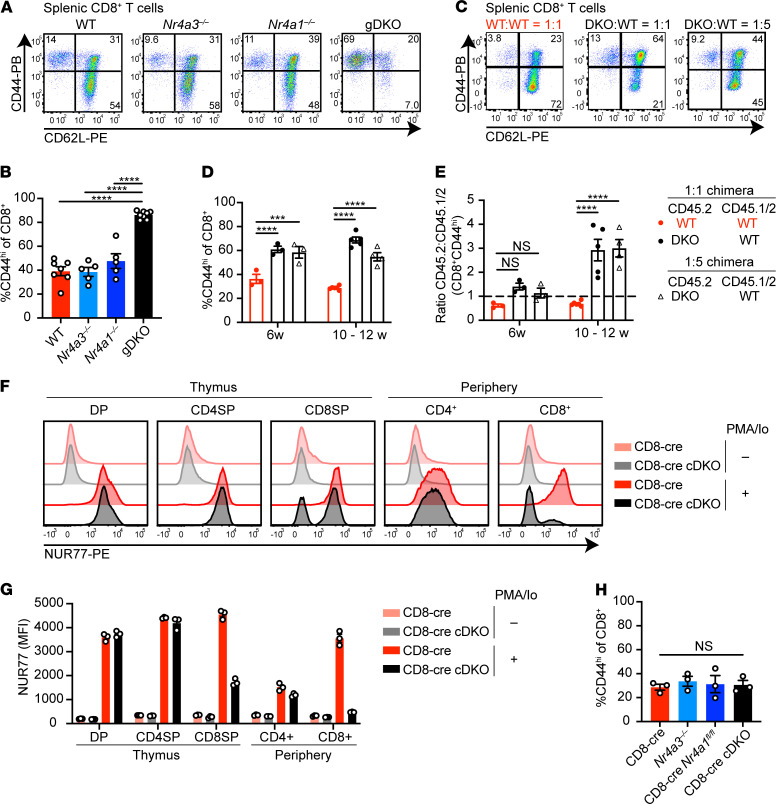
Figure 5. Reconstitution of WT Treg compartment does not restore CD8+ T cell homeostasis in competitive chimeras.
(A) Splenocytes from WT, Nr4a3–/–, Nr4a1–/–, and gDKO mice were stained to detect CD8+ T cell subsets on the basis of CD44 and CD62L expression. Plots are representative of ≥5 mice/genotype. (B) Quantification of splenic CD44hiCD8+ T cells as gated in A (n ≥ 5, 3- to 4-week-old gDKO and 5- to 6-week-old mice of other genotypes). (C) Flow plots showing the peripheral CD8+ T cell subsets in competitive chimeras, as described for A above. Representative of ≥7 chimeras of each type. (D) Quantification of splenic CD44hiCD8+ T cells from chimeras as gated in C at varied time points posttransplant (n ≥ 3). (E) Ratio of CD45.2 to CD45.1/2 for CD8+CD44hi population as gated in C, normalized to naive CD8+CD44loCD62Lhi gate (n ≥ 3). Data in C–E pooled from 2 sets of independently generated chimeras. (F and G) Thymocytes and splenocytes from CD8-cre and CD8-cre Nr4a1fl/fl Nr4a3–/– (cDKO) mice were stimulated with PMA and ionomycin (PMA/Io) for 2 hours. Flow plots show intracellular NUR77 expression following fixation and permeabilization within thymic and splenic T cell subsets (F). Quantification of NUR77 MFI in T cell subsets (G) (n = 3 mice/genotype). (H) Quantification of splenic CD8+CD44hi T cells from CD8-cre, Nr4a3–/–, CD8-cre Nr4a1fl/fl, and CD8-cre cDKO mice (n = 3 mice/genotype). Graphs depict mean ± SEM. Statistical significance was assessed by 1-way ANOVA with Tukey’s test (B and H) or 2-way ANOVA with Dunnett’s test (D and E). ***P < 0.001; ****P < 0.0001. NS, not significant.