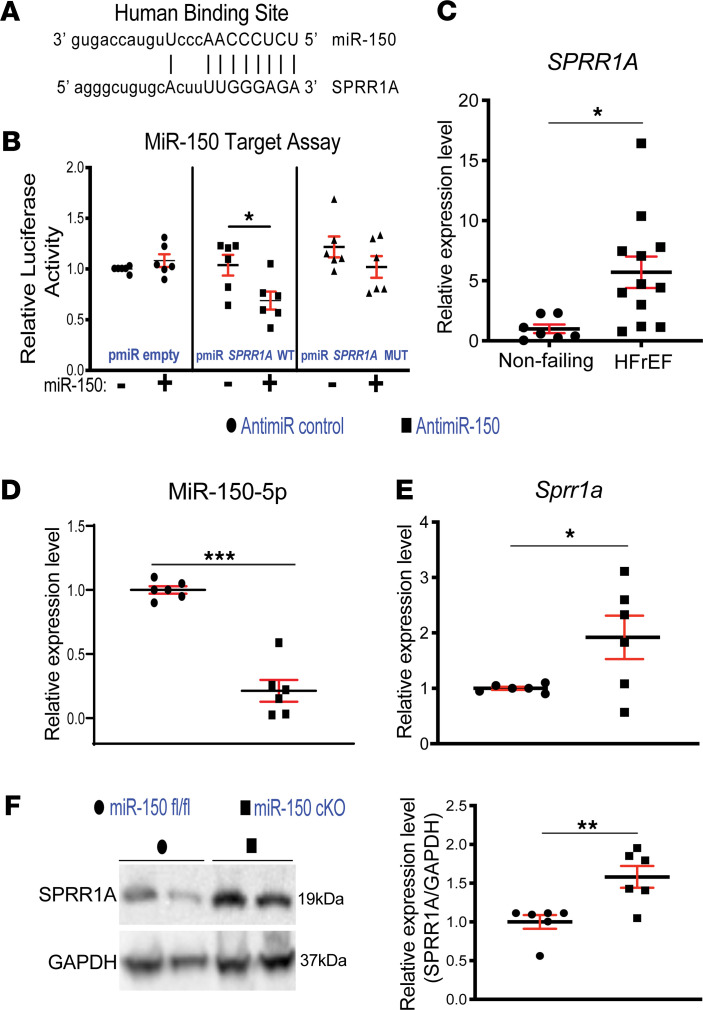
Figure 5. SPRR1A is a direct target of miR-150 and is upregulated in patients with heart failure.
(A) Human SPRR1A has a miR-150 binding site. miR-150 seed pairing in the target region is presented as vertical lines. (B) The ability of miR-150 to directly inhibit the activity of luciferase (LUC) reporter constructs that contain either WT or mutated (MUT) binding site for SPRR1A. Transfection with or without miR-150 mimic in H9c2 cells is shown. Firefly LUC activity was normalized to Renilla LUC activity and compared with empty vector measurements. Results are representative of 6 independent experiments with different biological samples. Unpaired 2-tailed t test. *P < 0.05 versus miR mimic control. (C) qPCR expression analysis of SPRR1A in heart tissues from patients with heart failure with reduced ejection fraction (HFrEF; n = 12) relative to nonfailing heart tissues (n = 7). Data are shown as fold change of gene expression normalized to GAPDH. Unpaired 2-tailed t test. *P < 0.05 versus nonfailing. (D and E) RNAs isolated from H9c2 cells transfected with 100 nM MirVana miR-150 inhibitor or 15-mer control were analyzed by miR-150–specific qPCR to access the levels of miR-150 (D). Levels of Sprr1a are indicated in E. Data were normalized to U6 snRNA (D) or Gapdh (E) and expressed relative to antimiR control. Results are from 6 independent experiments with different biological samples. Unpaired 2-tailed t test. *P < 0.05 or ***P < 0.001 versus antimiR control. (F) SPRR1A protein levels were measured in whole heart lysates from miR-150 cKO mice compared with miR-150fl/fl. Results are from 6 independent experiments with different biological samples. Unpaired 2-tailed t test. **P < 0.01 versus miR-150fl/fl. Data are presented as mean ± SEM.