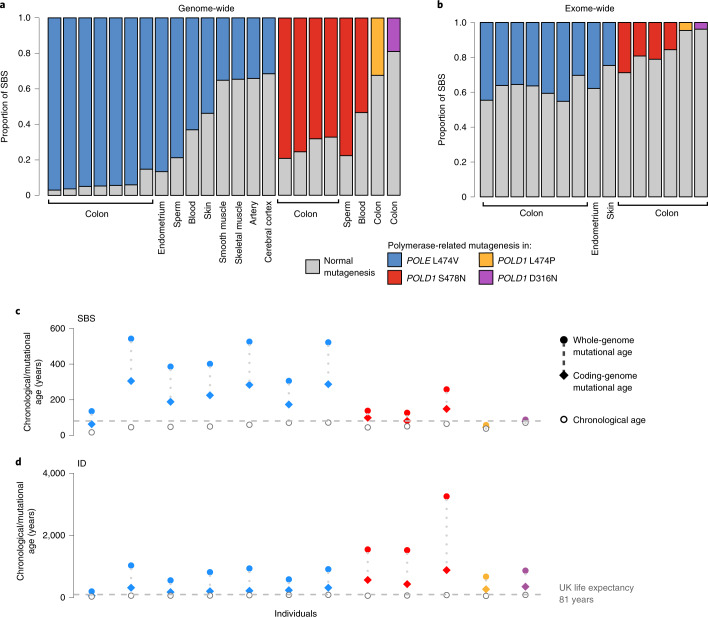
Fig. 4. Genome- and coding-sequence-wide increase in mutation burdens.
a, Genome-wide proportion of mutations due to POLE and POLD1 germline mutations across various normal tissues. Colored bars indicate mutagenesis due to POLE (SBS10a, SBS10b, SBS28) or POLD1 (SBS10c, SBS28) mutational signatures, with normal signatures in gray (SBS1, SBS5 and, for skin, SBS7a and SBS7b). b, Protein-coding exome proportion of mutations due to POLE and POLD1 germline mutations across various normal tissues, showing a much lower increase in polymerase-related mutational signatures (Wilcoxon signed-rank test P = 6.1 × 10−5). c,d, Mutational and chronological ages of histologically normal intestinal crypts per individual. Mutational ages are calculated based on the expected rate of mutation accumulation in wild-type intestinal crypts28, enabling the calculation of both SBS and ID mutational age. Plots show SBS (c) and ID (d) mutational ages across the whole genome (filled dots) and coding genome (filled diamonds). Germline mutation is color coded: blue, red, green and purple denote POLE L424V, POLD1 S478N, POLD1 L474P and POLD1 D316N, respectively. Individuals’ chronological ages are indicated by unfilled circles, and UK life expectancy is displayed as a dashed horizontal line.