Figure 6:
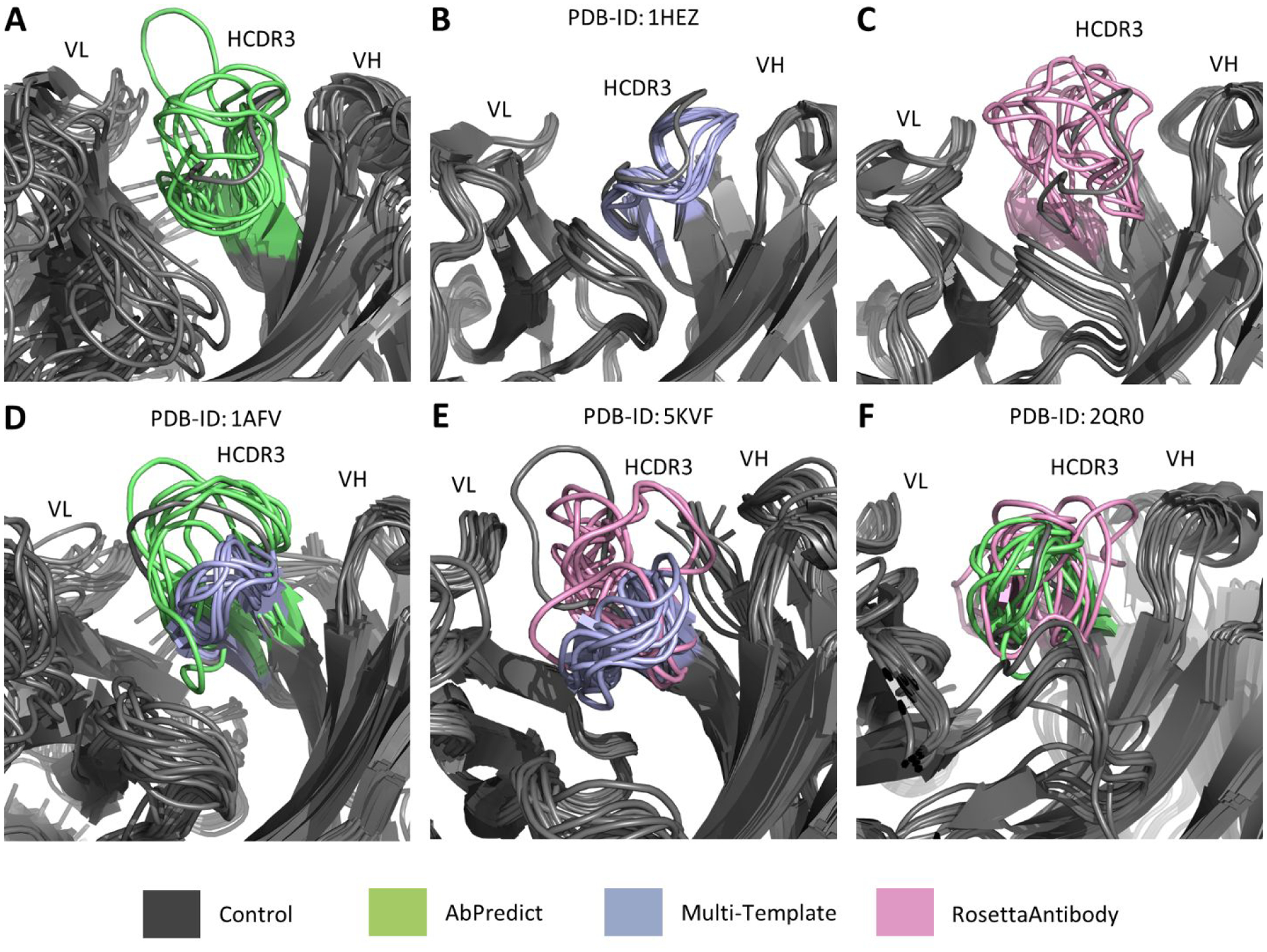
Examples of significant differences in HCDR3 modeling performance between multi-template modeling and AbPredict/RosettaAntibody. The top 10 models by score in Rosetta Energy Units for the two methods being compared in each example were aligned onto the corresponding antibody’s relaxed native crystal structure (grey). Our multi-template modeling method, displayed in purple (B), significantly outperformed AbPredict, displayed in green (A) and RosettaAntibody, displayed in pink (C), in modeling the HCDR3 loop of a Peptostreptococcus magnus protein L-binding human antibody (PDB ID 1HEZ)25. AbPredict only significantly outperformed our multi-template method in modeling the HCDR3 loop (D) when modeling an HIV-1 capsid protein-binding human antibody (PDB ID 1AFV)26. RosettaAntibody significantly outperformed our multi-template method in modeling the HCDR3 loop (E) when modeling the Zika virus envelope protein-binding human antibody (PDB ID 5KVF)27. AbPredict significantly outperformed RosettaAntibody in modeling the HCDR3 loop (F) when modeling a human vascular endothelial growth factor-binding antibody (PDB ID 2QR0)28.
