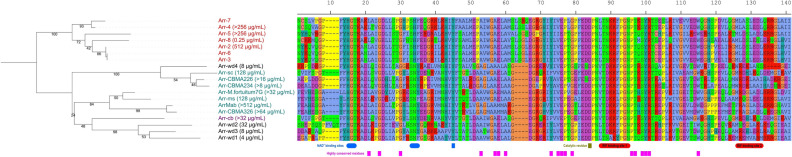
Figure 3.
Arr phylogenetic tree and sequence alignment. The colored labels indicate the phyla of the organisms from which the sequences were obtained: red, Proteobacteria; aqua, Actinobacteria; purple, Firmicutes; black, environment. The value in parentheses corresponds to the MIC given by Arr expressed in heterologous systems. Highlighted residues are based on the clustalX color scheme. Conserved residues and motifs are shown below the alignment, as follows: NAD+ binding sites, blue; catalytic residue, olive; RIF binding sites, red. Highly conserved residues (≥ 90%), considering all Arr sequences analyzed in the phylogeny, are represented below the alignment by the fuchsia blocks. References: Arr-2, Arr-sc, and Arr-ms1; Arr-4 and Arr-56; Arr-88; Arr-w1, Arr-w2, Arr-w3, and Arr-w44; ArrMab7; Arr-cb2.