Abstract
Small heat shock proteins (sHSPs) function as molecular chaperones in multiple physiological processes and are active during thermal stress. sHSP expression is controlled by heat shock transcription factor (HSF); however, few studies have been conducted on HSF in agricultural pests. Liriomyza trifolii is an introduced insect pest of horticultural and vegetable crops in China. In this study, the master regulator, HSF1, was cloned and characterized from L. trifolii, and the expression levels of HSF1 and five sHSPs were studied during heat stress. HSF1 expression in L. trifolii generally decreased with rising temperatures, whereas expression of the five sHSPs showed an increasing trend that correlated with elevated temperatures. All five sHSPs and HSF1 showed an upward trend in expression with exposure to 40 ℃ without a recovery period. When a recovery period was incorporated after thermal stress, the expression patterns of HSF1 and sHSPs in L. trifolii exposed to 40 °C was significantly lower than expression with no recovery period. To elucidate potential interactions between HSF1 and sHSPs, double-stranded RNA was synthesized to knock down HSF1 in L. trifolii by RNA interference. The knockdown of HSF1 by RNAi decreased the survival rate and expression of HSP19.5, HSP20.8, and HSP21.3 during high-temperature stress. This study expands our understanding of HSF1-regulated gene expression in L. trifolii exposed to heat stress.
Keywords: Liriomyza trifolii, HSF1, sHSPs, Heat stress, RNAi
Introduction
The leafminer fly, Liriomyza trifolii (Burgess), is an invasive, polyphagous pest that damages agricultural and horticultural crops worldwide (Spencer 1973). L. trifolii larvae form tunnels in foliage, and adults pierce leaf tissue for feeding and oviposition (Johnson et al. 1983; Parrella et al. 1985; Reitz et al. 1999). L. trifolii was first reported in mainland China in 2005 after the earlier invasion of L. sativae and L. huidobrensis (Wang et al. 2007). These three Liriomyza spp. are regarded as the most serious leafminer flies in the field (Wen et al. 1996; Kang et al. 2009; Xiang et al. 2012; Wan and Yang 2016), and multiple reports have documented interspecific competition among Liriomyza spp. (Gao et al. 2017). Because of its strong competitiveness, L. trifolii supplanted L. sativae in regions of southern China and has emerged as the predominant species (Wen et al. 1998; Wang et al. 2007; Wan and Yang 2016; Gao et al. 2017; Chang et al. 2017a, 2020a).
Temperature has played a major role in the development and distribution of Liriomyza spp. (Kang et al. 2009). Minor variations in thermotolerance were shown to perturb the competitive balance among congener species (Kang et al. 2009; Wang et al. 2014a, b). Several studies have investigated the response of Liriomyza spp. to temperature and thermally regulated interspecific competition (Reitz and Trumble 2002; Abe and Tokumaru 2008; Wang et al. 2014a, b). Genes encoding heat shock proteins (HSPs) have been identified and compared among three Liriomyza spp. (Huang and Kang 2007; Chang et al. 2017a). HSPs function as molecular chaperones (Gehring and Wehner 1995; Johnston et al. 1998), and their overproduction is a well-established indicator of insect tolerance to thermal stress (Feder and Hofmann 1999; Hu et al. 2014). Among HSPs, the small heat shock protein (sHSP) family has the largest diversity in function and structure and also exhibits thermoprotective properties (Gehring and Wehner 1995; Franck et al. 2004). In Chilo suppressalis, two sHSPs (CsHSP19.8 and CsHSP24.3) were upregulated by both low and high thermal stress (Lu et al. 2014; Pan et al. 2017). In other studies, expression levels of sHSP genes in the Eastern spruce budworm (Quan et al. 2017), western flower thrips (Wang et al. 2014a, b), and the oriental fruit fly (Dou et al. 2017) were significantly upregulated during temperature stress. Similarly, sHSPs in closely related Liriomyza spp. were induced by high- and low-temperature stress and exhibited different expression patterns (Huang and Kang 2007; Chang et al. 2019).
The expression of HSPs is controlled by the heat shock transcription factor (HSF) family (Wiederrecht et al. 1988). Within the typical four-membered HSF family, HSF1 is regarded as the master regulator (Wu 1995; Åkerfelt et al. 2010; Gomez-Pastor et al. 2018). HSF1 is activated by phosphorylation and trimer formation and then imported to the nucleus where it activates HSP expression (Neef et al. 2011; Steurer et al. 2018; Kovács et al. 2019). Specifically, HSF1 binds to the heat shock element (HSE) in the promoter region of target genes; the HSE contains at least three subsequent inverted repeats (e.g. TTCnnGAAnnTTC) (Amin et al. 1988). A single form of HSF (HSF1) is present in Drosophila melanogaster; however, other eukaryotes may contain multiple forms of HSF (Fujikake et al. 2005; Shamovsky and Nudler 2008; Fujimoto and Nakai 2010; Anckar and Sistonen 2011; Neudegger et al. 2016). Although HSF1 has been investigated in Drosophila and mammals, studies on HSF1 in agricultural insect pests are lacking.
In this study, HSF1 was cloned and characterized from L. trifolii, and the expression of HSF1 and five sHSPs was investigated during heat stress. HSF1 expression was silenced in L. trifolii using dsRNA to better understand the relationship between HSF1 and sHSPs. The results provide insight regarding the role of HSF1 in regulating sHSP expression during heat stress.
Materials and methods
Insects
L. trifolii was maintained at 25 ± 1 °C with a 16:8-h (light:dark) photoperiod as described previously (Chen and Kang 2002). L. trifolii was reared on kidney beans (Phaseolus vulgaris), and foliage with tunnels was collected for pupation and eclosion.
Cloning and sequence alignment of HSF1
Total RNA was isolated from L. trifolii with RNA-easy reagent (Vazyme, China, #R701), and RNA quality was assessed as described previously (Chang et al. 2019). A fragment of HSF1 was amplified using transcriptome data (Chang et al. 2020b) and verified with gene-specific primers (Table 1). The complete HSF1 cDNA was obtained with the SMART RACE cDNA Amplification Kit as described (Chang et al. 2019).
Table 1.
Primers used for cDNA cloning, dsRNA synthesis, and real-time quantitative PCR
| Gene | Primer sequences (5ʹ → 3ʹ) | Fragment length (bp) | |
|---|---|---|---|
| cDNA cloning and full-length cDNA amplification | |||
| HSF1 | F | ACCCAAAAACTGACCACCTTAT | 394 |
| R | ATGAGTGAAAAGCGTGAGTCCA | ||
| 5ʹ | GCGTGAGTCCAGTGAATCCTGCCTA | ||
| 3ʹ | GAGGCAGTGACTAAAGTGCTACAGG | ||
| dsRNA synthesis | |||
| dsHSF1 | F | TAATACGACTCACTATAGGGAGA TGGACTCACGCTTTTCACTC | 702 |
| R | TAATACGACTCACTATAGGGAGA ATAAAGACTCCTGACGCATC | ||
| dsGFP | F | TAATACGACTCACTATAGGGAGA CCTCGTGACCACCCTGACCTAC | 314 |
| R | TAATACGACTCACTATAGGGAGA CACCTTGATGCCGTTCTTCTGC | ||
| qRT-PCR | |||
| HSF1 | F | ATTGTCCCTACCTGTTGGAGCAC | 99 |
| R | GCACTTTAGTCACTGCCTCTGGT | ||
| HSP19.5 | F | GCTACCTGTTGCCTGAAAATGCT | 118 |
| R | CTTTGGTTCATCTTTTGGTGCTG | ||
| HSP20.8 | F | AACAACTGGTGGGATGACTATGAC | 144 |
| R | CATTTTGTGTCTGCTGCTGTTGT | ||
| HSP21.3 | F | GAAATCAATGTGAAAGTGGTGGA | 175 |
| R | GAACCTTCAACAAGCCATCAGAT | ||
| HSP21.7 | F | CAACAGTTTGCTCCCAATGAAG | 125 |
| R | GAGGTAGCGTCTGGAGAAGTGA | ||
| HSP21.7b | F | CTCCAGACGCTACCTCTTGCCT | 159 |
| R | CTTAGCAGGTTGATTGGTTTGAGT | ||
| ACTIN | F | TTGTATTGGACTCTGGTGACGG | 73 |
| R | GATAGCGTGAGGCAAAGCATAA | ||
Note: F, forward; R, reverse; 5ʹ, 5ʹ RACE primer; 3ʹ, 3ʹ RACE primer; underscored nucleotides indicate the T7 polymerase promoter sequence
The HSF1 cDNA sequence was used in BLAST searches (http://www.ncbi.nlm.gov/BLAST/) to identify homologs in other insect species. Clustal X (Thompson et al. 1997) and ORF Finder (https://www.ncbi.nlm.nih.gov/orffinder/) were used to align sequences and identify ORFs, respectively. The ExPASy Molecular Biology Server and MEGA 6.0 (Tamura et al. 2013) were used to predict HSF1 protein sequences and phylogenetic relationships with the neighbor-joining method as described previously (Chang et al. 2019). The 3D structure of the DBD domain was predicted by the SWISS-MODEL website (https://swissmodel.expasy.org/) using the D. melanogaster DBD domain (SMTL ID: 1hkt.1) as a template.
High-temperature treatment and expression of HSF1 and sHSPs
Newly emerged L. trifolii adults (n = 10) were collected and exposed to 35, 37.5, 40, 42.5, and 45 °C for 1 h in a water bath (DC-3010, Ningbo, China). Adults maintained at 25 °C served as controls. Following exposure to high temperatures, L. trifolii were allowed to recover at 25 °C for 1 h and were then frozen in liquid nitrogen and stored at − 80 °C. Treatments were repeated three times. For different treatment and recovery time experiments, newly emerged adults (n = 10; six biological replicates per temperature) were exposed for 15, 30, 60, and 120 min at 40 °C. Control adults were maintained at 25 °C. Following exposure, half of the samples (three biological replicates) were allowed to recover at 25 °C for the same length of time as the treatment (e.g. 15, 30, 60, and 120 min) and the other half (three biological replicates) were immediately frozen in liquid nitrogen, and stored at − 80 °C for RNA extraction.
Reverse transcription of total RNA (0.5 μg) was performed with the Bio-Rad iScript™ cDNA Synthesis Kit (Bio-Rad, CA, USA). qRT-PCR reactions were performed as described (Chang et al. 2017b) with a CFX-96 real-time PCR system (Bio-Rad). Gene-specific primers are shown in Table 1. Treatments contained four biological replicates, and each reaction was performed in triplicate.
dsRNA synthesis and RNAi
The full-length L. trifolii HSF1 was analyzed with siDirect v. 2.0 (http://sidirect2.rnai.jp/) to select potential small interfering RNA (siRNA) sequences that could be used to design dsRNA primers. A T7 promoter sequence (TAATACGACTCACTATAGGGAGA) was incorporated into the 5ʹ end of sense and antisense primers to facilitate transcription from both cDNA strands. The control consisted of dsRNA specific to green fluorescence protein (GFP) (Table 1). Purified DNA template (1.5 µg) was used to synthesize dsRNA, and products were purified with the MEGAscript™ RNAi Kit (Thermo, USA, #AM1626). The quality and quantity of dsRNA were evaluated by gel electrophoresis and spectrophotometry, respectively.
After anesthesia with CO2, newly emerged L. trifolii adults were microinjected with dsRNA (Nanoliter 2010 Injector, WPI, USA). Insects were microinjected with 5 nL (total 50 ng) of dsGFP or dsHSF1 and caged on a modified 96-well plate containing a honey/water solution throughout the experiment; dead insects were removed as needed. In silencing efficiency experiments, 50 ng of dsHSF1 was injected per adult, and insects were subjected to 40 °C for 30 min. Silencing efficiency in surviving adults was measured at 24 h post-injection by qRT-PCR. Each treatment was repeated three times. Survival rates were also calculated in treatments containing 10 injected adults; the number of viable and expired adults was recorded after exposure to 40 °C for 30 min. Each treatment was repeated seven times.
Statistical analyses
Expression levels of HSF1 and sHSP genes at different temperatures and exposure times were calculated using the 2−ΔΔCt method (Livak and Schmittgen 2001), and ACTIN served as a reference gene (Chang et al. 2017b). One-way ANOVA (Tukey’s multiple comparison) was used to detect significant differences among temperatures using SPSS v. 16.0 (SPSS, Chicago, IL, USA). For ANOVA, data were transformed for homogeneity of variances, and differences were considered statistically significant when P < 0.05.
For silencing efficiency, the relative abundance of target genes and survival rates were compared to the dsGFP control. Student’s t-test was used to compare differences in gene expression and mortality with SPSS v. 16.0, and differences were considered significant at P < 0.05.
Results
Characterization and phylogenetic analysis of L. trifolii HSF1
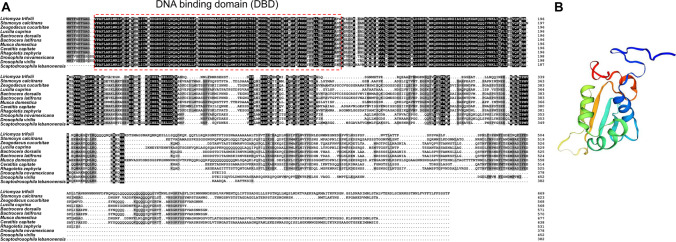
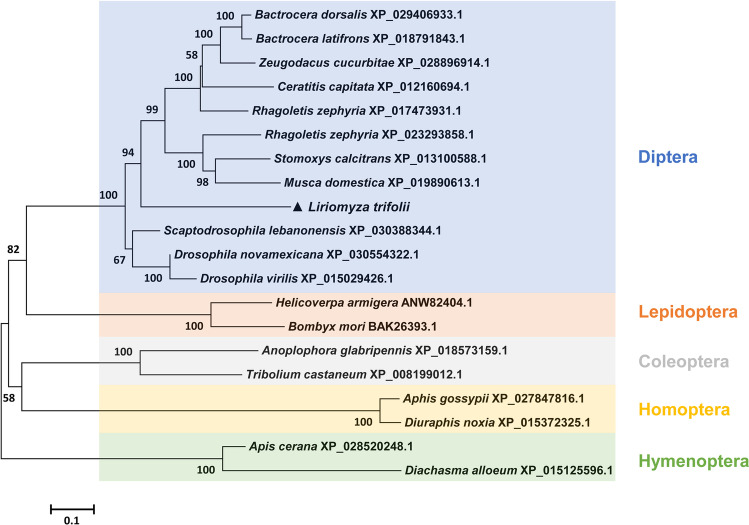
HSF1 was cloned from L. trifolii and deposited in GenBank (accession no. MW054859); the 2013-bp ORF encodes a 75.84-kDa protein with 670 amino acids. The deduced L. trifolii HSF1 protein sequence contained a typical DNA-binding domain (DBD) (Fig. 1(A)). The three-dimensional (3D) structure of HSF1 from L. trifolii was modeled using the DBD domain in D. melanogaster (SMTL ID: 1hkt.1) as a template (Fig. 1(B)); HSF1 showed 83.02% sequence identity to the D. melanogaster orthologue. To examine relationships between HSF1s, a phylogenetic tree was generated using the neighbor-joining method; this included 12 Dipteran species and eight other insect species. HSF1 in L. trifolii clustered with HSF1 orthologues in other members of Diptera (Fig. 2).
Fig. 1.
Structure of HSF1 and multiple sequence alignment of HSF1 from various insect species. (A) Sequence alignment of predicted HSF1 proteins in various insect species. (B) 3D structure of HSF1 in L. trifolii. The deduced HSF1 protein contained a DNA-binding domain (DBD), and the 3D structure using the D. melanogaster DBD domain was used as a template
Fig. 2.
Phylogenetic analysis of HSF1 from L. trifolii and other insects. The neighbor-joining algorithm was used to construct the tree; numbers on branches are bootstrap values obtained from 1,000 replicates (only bootstrap values > 50 are shown). HSF1 from L. trifolii is marked with a triangle
Expression of HSF1 and sHSPs in L. trifolii adults during heat stress
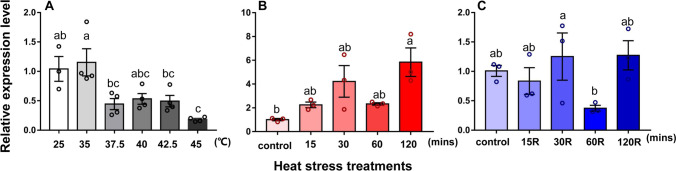
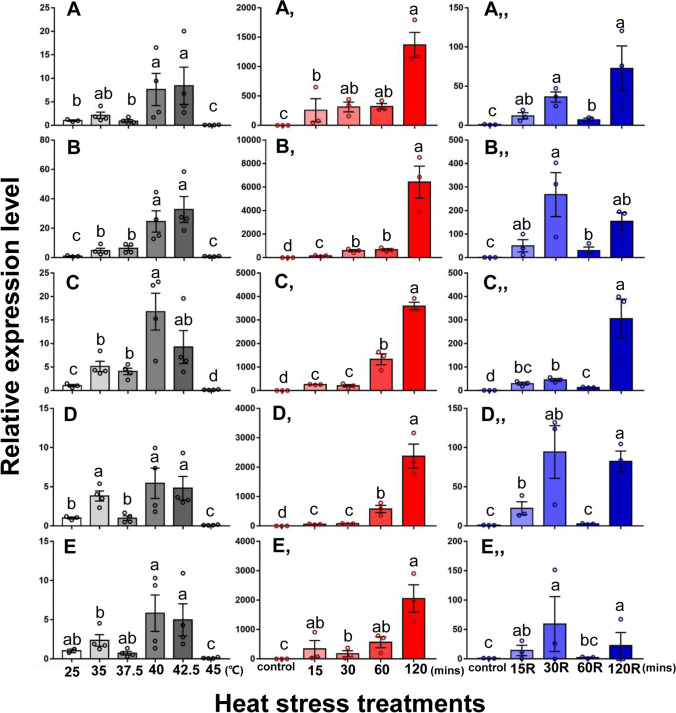
Expression levels of HSF1 and five sHSPs (HSP19.5, HSP20.8, HSP21.3, HSP21.7, and HSP21.7b) were evaluated during heat stress. Compared to the control group at 25 °C, the expression levels of HSF1 were significantly lower at 35 to 45 °C (F5,17 = 7.345, P < 0.001) (Fig. 3(A)). However, expression levels of the five sHSPs were significantly increased at elevated temperatures (HSP19.5: F5,17 = 19.423, P < 0.001; HSP20.8: F5,17 = 36.641, P < 0.001; HSP21.3: F5,17 = 44.554, P < 0.001; HSP21.7: F5,17 = 29.235, P < 0.001; HSP21.7b: F5,16 = 15.980, P < 0.001) (Fig. 4(A–E)). The highest expression of sHSPs occurred at 40 or 42.5 °C, and these expression levels were much higher than the control at 25 °C (Fig. 4(A–E)).
Fig. 3.
Relative expression levels of HSF1 from L. trifolii during heat stress. (A) Expression during different periods of high-temperature stress. (B) Expression during different periods of heat stress with no recovery period. (C) Expression during different periods of heat stress with recovery. Control adults were maintained at 25 °C. Different lowercase letters indicate significant differences between treatments. Tukey’s multiple range test was used for pairwise comparison of means (P < 0.05)
Fig. 4.
Relative expression of five LtHSPs during different heat stress. Expression of (A) LtHSP19.5, (B) LtHSP20.8, (C) LtHSP21.3, (D) LtHSP21.7, and (E) LtHSP21.7b at different temperatures. Expression levels of (Aʹ) LtHSP19.5, (Bʹ) LtHSP20.8, (Cʹ) LtHSP21.3, (Dʹ) LtHSP21.7, and (Eʹ) LtHSP21.7b during periods of high-temperature stress with no recovery period. Expression levels of (Aʺ) LtHSP19.5, (Bʺ) LtHSP20.8, (Cʺ) LtHSP21.3, (Dʺ) LtHSP21.7, and (Eʺ) LtHSP21.7b under different periods of heat stress with recovery. Control adults were maintained at 25 °C. Different lowercase letters indicate significant differences among treatments. Tukey’s multiple range test was used for pairwise comparison of means (P < 0.05)
In experiments where L. trifolii was exposed to 40 °C for 15–120 min without a recovery period, expression of HSF1 was significantly higher at 120 min vs. the control (F4,10 = 10.808, P < 0.05) (Fig. 3(B)). When a recovery period was incorporated into the experiment, expression of HSF1 was significantly lower at 60 vs. 30 min (F4,10 = 3.7288, P < 0.05), but there was no significant difference among other treatment times (Fig. 3(C)).
Expression of sHSPs was also evaluated in L. trifolii adults exposed to 40 °C without a recovery period; expression levels for all five sHSPs showed significant increases beginning at 15 min and were highest at 120 min (Fig. 4(Aʹ–Eʹ)) (HSP19.5: F4,10 = 44.520, P < 0.001; HSP20.8: F4,10 = 395.162, P < 0.001; HSP21.3: F4,10 = 382.432, P < 0.001; HSP21.7: F4,10 = 347.760, P < 0.001; HSP21.7b: F4,10 = 22.197, P < 0.001). When a recovery period was incorporated into the experiment, the expression of the five sHSPs increased at different treatment times (HSP19.5: F4,10 = 19.356, P < 0.001; HSP20.8: F4,10 = 23.762, P < 0.001; HSP21.3: F4,10 = 103.307, P < 0.001; HSP21.7: F4,10 = 45.154, P < 0.001; HSP21.7b: F4,10 = 11.365, P < 0.001). The five sHSPs showed a consistent spike in expression levels at 30 and 120 min (Fig. 4(Aʺ–Eʺ)). Interestingly, there were no significant differences for HSP21.7 and HSP21.7b expression in the 60 min treatment (Fig. 4(Dʺ–Eʺ)).
Silencing HSF1 leads to decreased heat stress tolerance in L. trifolii
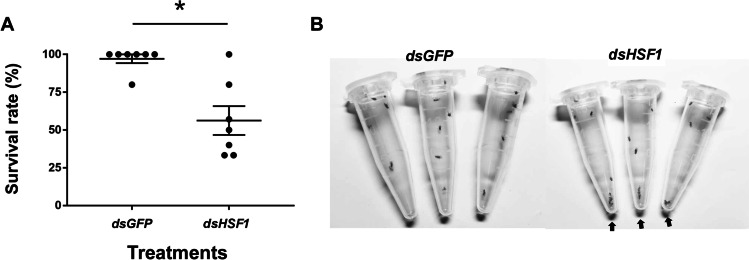
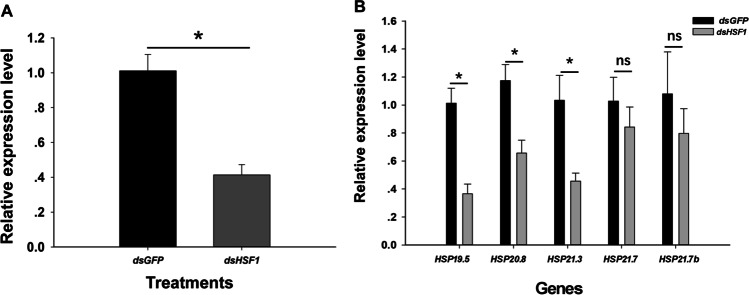
When insects were exposed to 40 °C for 30 min, survival was significantly reduced (~ 56.26%) in L. trifolii injected with dsHSF1 as compared to dsGFP (Fig. 5). Quantification of mRNA by qRT-PCR revealed that HSF1 expression was diminished by 58.65% in comparison to the dsGFP control (t = 5.430, P < 0.05) (Fig. 6(A)).
Fig. 5.
RNAi-mediated knockdown of HSF1 decreases L. trifolii survival. (A) Percentage of survival in insects injected with dsHSF1 and dsGFP (control). (B) Insect viability after RNAi; arrows represent dead insects. Data were analyzed by Student’s t-test, P < 0.05. Asterisks represent significant differences in survival of insects injected with dsHSF1 and the dsGFP control
Fig. 6.
HSF1 and sHSP expressions in L. trifolii injected with dsHSF1 and dsGFP. (A) Expression of HSF1. (B) Expression of LtHSP19.5, LtHSP20.8, LtHSP21.3, LtHSP21.7, and LtHSP21.7b. Data were analyzed by Student’s t-test, P < 0.05. Asterisks represent significant differences between dsGFP and dsHSF1-treated insects; ns indicates no significant difference
L. trifolii treated with dsHSF1 were significantly impaired in transcription of HSP19.5, HSP20.8, and HSP21.3, which were reduced by 63.50, 34.35, and 54.36% as compared to dsGFP-injected controls (HSP19.5: t = 5.068, P < 0.05; HSP20.8: t = 3.515, P < 0.05; HSP21.3: t = 3.082, P < 0.05) (Fig. 6(B)). There were no significant differences in the expression of HSP21.7 and HSP21.7b in dsHSF1- and dsGFP-treated flies (HSP21.7: t = 0.836, P = 0.450; HSP21.7b: t = 0.813, P = 0.462) (Fig. 6(B)).
Discussion
The deduced protein sequence of HSF1 from L. trifolii contained the conserved DBD region, which binds to HSEs in target genes (Neudegger et al. 2016). HSF proteins use the DBD region to activate HSPs and other genes that encode molecular chaperones (Brunquell et al. 2016; Mahat et al. 2016; Li et al. 2017; Takii et al. 2017). The deduced 3D structures of HSF1 share typical features of the HSF1 family and phylogenetic analysis of HSF1 from L. trifolii revealed close relatedness with orthologous proteins in Diptera, which indicates conservation in the HSF1 family.
In general, expression of HSF1 in L. trifolii decreased with rising temperatures, whereas expression of the five sHSPs showed an increasing trend that was concomitant with rising temperatures. These opposing transcription patterns are indicative of the temporal nature of gene expression in response to heat stress. When HSF is exposed to temperature stress, it shows a rapid increase in transcription, which is followed by the induction of sHSPs. When sHSP transcription is induced, HSF1 is gradually downregulated to allow for more HSP expression (Nielsen et al. 2005; Anckar and Sistonen 2007). In this study, the five sHSPs showed varied responses to temperature, which is indicative of the cooperative effect of sHSP family members with respect to thermotolerance. In a previous study, expression of sHSPs in L. trifolii pupae showed diverse patterns during heat stress, which indicates the underlying complexity in sHSP evolution (Chang et al. 2019).
HSF1 was highly expressed in the 30-min recovery and 120-min “no recovery” treatments, which supports the contention that HSF1 responds rapidly to thermal stress. However, it should be noted that the overall expression levels in treatments without recovery were higher than those in the recovery treatment. Furthermore, expression of the five sHSPs was generally higher at 30 and 120 min with a recovery period (Fig. 4(Aʺ–Eʺ)), which is a pattern similar to HSF1. In Litopenaeus vannamei, HSP70 was upregulated when HSF1 was overexpressed, which suggested that HSF1 might function to activate the HSP70 promoter (Yan et al. 2014). The two peaks in expression at 30 and 120 min after recovery suggest that other mechanisms of high-temperature tolerance are active. In Frankliniella occidentalis, FoHSP706 expression varied at different developmental stages and treatment times (Zhang et al. 2019). In soybean seedlings, HSPs accumulated within minutes after heat shock and peaked at 1–2 h (Kimpel et al. 1990). In D. melanogaster, the expression of DmHSP70Aa peaked at 2 h following recovery after exposure to 0 °C (Colinet et al. 2010).
Suppression of HSF1 by RNAi decreased survival and sHSP expression in L. trifolii exposed to thermal stress. The high rate of mortality indicates that HSF1 plays a key role in thermotolerance; however, there are likely other mechanisms in place to ensure that L. trifolii can survive thermal stress (Guertin et al. 2010; Chang et al. 2020b). Our results show that suppression of HSF1 decreased HSP19.5, HSP20.8, and HSP21.3 expression, but had no significant effect on transcription of HSP21.7 and HSP21.7b. These data may indicate that HSF1 binds to HSEs in HSP19.5, HSP20.8, and HSP21.3 to regulate their expression, while HSP21.7 and HSP21.7b may be regulated by other transcription factors. Interestingly, HSP21.7 and HSP21.7b expression levels were not as high as the other three sHSPs during temperature stress. In Artemia franciscana, HSF1 knockdown led to a reduction in stress tolerance proteins in diapausing embryos, which suggests that HSF1 regulates stress-related genes (Tan and Macrae 2018). In the flea beetle, Agasicles hygrophila, injection of newly emerged adults with dsAhHSF reduced the transcription of two HSPs and caused reductions in egg production and survival (Jin et al. 2020). These findings indicate that further studies are needed to confirm interactions between HSF1 and sHSPs.
In summary, HSF1 was cloned from L. trifolii and expressions of HSF1 and five sHSPs were analyzed in response to different temperatures. To better understand the regulation of sHSPs expression, dsHSF1 was generated and used to silence HSF1 in L. trifolii. The knockdown of HSF1 increased mortality in L. trifolii adults during heat stress and reduced stress tolerance based on the expression levels of sHSPs. The findings indicate that HSF1 regulates the expression of stress-related genes when L. trifolii is exposed to high temperatures.
Acknowledgements
This research was funded by the earmarked fund for Jiangsu Agricultural Industry Technology System (JATS [2020] 309), the Jiangsu Science & Technology Support Program (BE2014410), and the postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX18_2374).
Declarations
Competing interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Abe Y, Tokumaru S. Displacement in two invasive species of leafminer fly in different localities. Biol Invasions. 2008;10:951–953. doi: 10.1007/s10530-007-9173-2. [DOI] [Google Scholar]
- Åkerfelt M, Morimoto RI, Sistonen L. Heat shock factors: integrators of cell stress, development and lifespan. Nat Rev Mol Cell Biol. 2010;8:545–555. doi: 10.1038/nrm2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin J, Ananthan J, Voellmy R. Key features of heat shock regulatory elements. Mol Cell Biol. 1988;8:3761–3769. doi: 10.1128/mcb.8.9.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anckar J, Sistonen L. Heat shock factor 1 as a coordinator of stress and developmental pathways. Adv Exp Med Biol. 2007;594:78–88. doi: 10.1007/978-0-387-39975-1_8. [DOI] [PubMed] [Google Scholar]
- Anckar J, Sistonen L. Regulation of HSF1 function in the heat shock response: implications in aging and disease. Annu Rev Biochem. 2011;80:1089–1115. doi: 10.1146/annurev-biochem-060809-095203. [DOI] [PubMed] [Google Scholar]
- Brunquell J, Morris S, Lu Y, Cheng F, Westerheide SD. The genome-wide role of HSF-1 in the regulation of gene expression in Caenorhabditis elegans. BMC Genomics. 2016;17:559. doi: 10.1186/s12864-016-2837-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YW, Chen JY, Lu MX, Gao Y, Tian ZH, Gong WR, Dong CS, Du YZ. Cloning and expression of genes encoding heat shock proteins in Liriomyza trifolii and comparison with two congener leafminer species. PLoS ONE. 2017;12:e0181355. doi: 10.1371/journal.pone.0181355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YW, Chen JY, Lu MX, Gao Y, Tian ZH, Gong WR, Zhu W, Du YZ. Selection and validation of reference genes for quantitative real time PCR analysis under different experimental conditions in the leafminer Liriomyza trifolii (Diptera: Agromyzidae) PLoS ONE. 2017;12:e0181862. doi: 10.1371/journal.pone.0181862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YW, Zhang XX, Lu MX, Du YZ, Zhu-Salzman K. Molecular cloning and characterization of small heat shock protein genes in the invasive leaf miner fly, Liriomyza TrifoliI. Genes. 2019;10:775. doi: 10.3390/genes10100775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YW, Wang YC, Zhang XX, Iqbal J, Lu MX, Gong HX, Du YZ. Comparative transcriptome analysis of three invasive leafminer flies provides insights into interspecific competition. Int J Biol Macromol. 2020;165:1664–1674. doi: 10.1016/j.ijbiomac.2020.09.260. [DOI] [PubMed] [Google Scholar]
- Chang YW, Zhang XX, Lu MX, Gong WR, Du YZ. Transcriptome analysis of Liriomyza trifolii (Diptera: Agromyzidae) in response to temperature stress. Comp Biochem Phys D. 2020;34:100677. doi: 10.1016/j.cbd.2020.100677. [DOI] [PubMed] [Google Scholar]
- Chen B, Kang L. Cold hardiness and supercooling capacity in the pea leafminer Liriomyza huidobrensis. Cryo Lett. 2002;23:173–182. [PubMed] [Google Scholar]
- Colinet H, Lee SF, Hoffmann A. Temporal expression of heat shock genes during cold stress and recovery from chill coma in adult Drosophila melanogaster. Febs J. 2010;277:174–185. doi: 10.1111/j.1742-4658.2009.07470.x. [DOI] [PubMed] [Google Scholar]
- Dou W, Tian Y, Liu H, Shi Y, Smagghe G, Wang JJ. Characteristics of six small heat shock protein genes from Bactrocera dorsalis: diverse expression under conditions of thermal stress and normal growth. Comp Biochem Physiol B Biochem Mol Biol. 2017;213:8–16. doi: 10.1016/j.cbpb.2017.07.005. [DOI] [PubMed] [Google Scholar]
- Feder ME, Hofmann GE. Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu Rev Physiol. 1999;61:243–282. doi: 10.1146/annurev.physiol.61.1.243. [DOI] [PubMed] [Google Scholar]
- Franck E, Madsen O, Van-Rheede T, Ricard G, Huynen MA, De-Jong WW. Evolutionary diversity of vertebrate small heat shock proteins. J Mol Evol. 2004;59:792–805. doi: 10.1007/s00239-004-0013-z. [DOI] [PubMed] [Google Scholar]
- Fujikake N, Nagai Y, Popiel HA, Kano H, Yamaguchi M, Toda T. Alternative splicing regulates the transcriptional activity of Drosophila heat shock transcription factor in response to heat/cold stress. FEBS Lett. 2005;17:3842–3848. doi: 10.1016/j.febslet.2005.05.074. [DOI] [PubMed] [Google Scholar]
- Fujimoto M, Nakai A. The heat shock factor family and adaptation to proteotoxic stress. FEBS J. 2010;277:4112–4125. doi: 10.1111/j.1742-4658.2010.07827.x. [DOI] [PubMed] [Google Scholar]
- Gao YL, Reitz SR, Xing ZL, Ferguson S, Lei ZR. A decade of a leafminer invasion in China: lessons learned. Pest Manag Sci. 2017;73:1775–1779. doi: 10.1002/ps.4591. [DOI] [PubMed] [Google Scholar]
- Gehring WJ, Wehner R. Heat shock protein synthesis and thermotolerance in Cataglyphis, an ant from the Sahara Desert. Proc Natl Acad Sci U S A. 1995;92:2994–2998. doi: 10.1073/pnas.92.7.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Pastor R, Burchfiel ET, Thiele DJ. Regulation of heat shock transcription factors and their roles in physiology and disease. Nat Rev Mol Cell Biol. 2018;19:4–20. doi: 10.1038/nrm.2017.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin MJ, Petesch SJ, Zobeck KL, Min IM, Lis JT. Drosophila heat shock system as a general model to investigate transcriptional regulation. Cold Spring Harb Symp Quant Biol. 2010;75:1–9. doi: 10.1101/sqb.2010.75.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JT, Chen B, Li ZH. Thermal plasticity is related to the hardening response of heat shock protein expression in two Bactrocera fruit flies. J Insect Physiol. 2014;67:105–113. doi: 10.1016/j.jinsphys.2014.06.009. [DOI] [PubMed] [Google Scholar]
- Huang LH, Kang L. Cloning and interspecific altered expression of heat shock protein genes in two leafminer species in response to thermal stress, Insect Mol. Biol. 2007;16:491–500. doi: 10.1111/j.1365-2583.2007.00744.x. [DOI] [PubMed] [Google Scholar]
- Jin JS, Li YZ, Zhou ZS, Zhang H, Guo JY, Wan FH. Heat shock factor is involved in regulating the transcriptional expression of two potential hsps (AhHsp70 and AhsHsp21) and its role in heat shock response of Agasicles hygrophila. Front Physiol. 2020;11:562204. doi: 10.3389/fphys.2020.562204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MW, Welter SC, Toscano NC, Tingi P, Trumble JT. Reduction of tomato leaflet photosynthesis rates by mining activity of Liriomyza sativae (Diptera: Agromyzidae) J Econ Entomol. 1983;76:1061–1063. doi: 10.1093/jee/76.5.1061. [DOI] [Google Scholar]
- Johnston JA, Ward CL, Kopito RR. Aggresomes: a cellular response to misfolded proteins. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang L, Chen B, Wei JN, Liu TX. Roles of thermal adaptation and chemical ecology in Liriomyza distribution and control. Annu Rev Entomol. 2009;54:127–145. doi: 10.1146/annurev.ento.54.110807.090507. [DOI] [PubMed] [Google Scholar]
- Kimpel JA, Nagao RT, Goekjian V, Key JL. Regulation of the heat shock response in soybean seedlings. Plant Physiol. 1990;94:988–995. doi: 10.1104/pp.94.3.988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovács D, Sigmond T, Hotzi B, Bohár B, Fazekas D, Deák V, Vellai T, Barna J. Hsf1base: a comprehensive database of hsf1 (heat shock factor 1) target genes. Int J Mol Sci. 2019;20:5815. doi: 10.3390/ijms20225815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Labbadia J, Morimoto RI. Rethinking HSF1 in stress, development, and organismal health. Trends Cell Biol. 2017;27:895–905. doi: 10.1016/j.tcb.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu MX, Hua J, Cui YD, Du YZ. Five small heat shock protein genes from Chilo suppressalis: characteristics of gene, genomic organization, structural analysis, and transcription profiles. Cell Stress Chaperon. 2014;19:91–104. doi: 10.1007/s12192-013-0437-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahat DB, Salamanca HH, Duarte FM, Danko CG, Lis JT. Mammalian heat shock response and mechanisms underlying its genome-wide transcriptional regulation. Mol Cell. 2016;62:63–78. doi: 10.1016/j.molcel.2016.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neef DW, Jaeger AM, Thiele DJ. Heat shock transcription factor 1 as a therapeutic target in neurodegenerative diseases. Nat Rev Drug Discov. 2011;10:930–944. doi: 10.1038/nrd3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neudegger T, Verghese J, Hayer-Hartl M, Hartl FU, Bracher A. Structure of human heat-shock transcription factor 1 in complex with DNA. Nat Struct Mol Biol. 2016;1:140–146. doi: 10.1038/nsmb.3149. [DOI] [PubMed] [Google Scholar]
- Nielsen MM, Overgaard J, Sørensen JG, Holmstrup M, Justesen J, Loeschcke V. Role of HSF activation for resistance to heat, cold and high-temperature knock-down. J Insect Physiol. 2005;51:1320–1329. doi: 10.1016/j.jinsphys.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Pan DD, Lu MX, Li QY, Du YZ. Characteristics and expression of genes encoding two small heat shock protein genes lacking introns from Chilo suppressalis. Cell Stress Chaperon. 2017;23:1–10. doi: 10.1007/s12192-017-0823-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrella MP, Jones VP, Youngman RR, Lebeck LM. Effect of leaf mining and leaf stippling of Liriomyza spp. on photosynthetic rates of chrysanthemum. Ann Entomol Soc A. 1985;78:90–93. doi: 10.1093/aesa/78.1.90. [DOI] [Google Scholar]
- Quan GX, Duan J, Ladd T, Krell PJ. Identification and expression analysis of multiple small heat shock protein genes in spruce budworm, Choristoneura fumiferana, (l.) Cell Stress Chaperon. 2017;23:141–154. doi: 10.1007/s12192-017-0832-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitz SR, Trumble JT. Interspecific and intraspecific differences in two Liriomyza leafminer species in California. Entomol Exp Appl. 2002;102:101–113. doi: 10.1046/j.1570-7458.2002.00930.x. [DOI] [Google Scholar]
- Reitz SR, Kund GS, Carson WG, Phillips PA, Trumble JT. Economics of reducing insecticide use on celery through low-input pest management strategies. Agric Ecosyst Environ. 1999;73:185–197. doi: 10.1016/S0167-8809(99)00016-X. [DOI] [Google Scholar]
- Shamovsky I, Nudler E. New insights into the mechanism of heat shock response activation. Cell Mol Life Sci. 2008;65:855–861. doi: 10.1007/s00018-008-7458-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer KA. Agromyzidae (Diptera) of economic importance. 9: Series Entomologica. Bath: The Hague Publishers; 1973. pp. 19–28. [Google Scholar]
- Steurer C, Eder N, Kerschbaum S, Wegrostek C, Gabriel S, Pardo N, Ortner V, Czerny T, Riegel E. Hsf1 mediated stress response of heavy metals. PLoS ONE. 2018;13:e0209077. doi: 10.1371/journal.pone.0209077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takii R, Fujimoto M, Matsuura Y, Wu F, Oshibe N, Takaki E, Katiyar A, Akashi H, Makino T, Kawata M, Naka A. HSF1 and HSF3 cooperatively regulate the heat shock response in lizards. PLoS ONE. 2017;12:e0180776. doi: 10.1371/journal.pone.0180776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan JB, Macrae TH. Stress tolerance in diapausing embryos of Artemia franciscana is dependent on heat shock factor 1 (Hsf1) PLoS ONE. 2018;13:e0200153. doi: 10.1371/journal.pone.0200153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan FH, Yang NW. Invasion and management of agricultural alien insects in China. Annu Rev Entomol. 2016;61:77–98. doi: 10.1146/annurev-ento-010715-023916. [DOI] [PubMed] [Google Scholar]
- Wang ZG, Guan W, Chen DH. Preliminary report of the Liriomyza trifolii in Zhongshan area. Plant Quarantine. 2007;21:19–20. [Google Scholar]
- Wang HH, Reitz SR, Xiang JC, Smagghe G, Lei ZR. Does Temperature-mediated reproductive success drive the direction of species displacement in two invasive species of leafminer fly? PLoS ONE. 2014;9:e98761. doi: 10.1371/journal.pone.0098761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HH, Rreitz S, Wang LX, Wang SY, Xue LI, Lei ZR. The mRNA expression profiles of five heat shock protein genes from Frankliniella occidentalis at different stages and their responses to temperatures and insecticides. J Integr Agric. 2014;13:2196–2210. doi: 10.1016/S2095-3119(13)60680-2. [DOI] [Google Scholar]
- Wen JZ, Wang Y, Lei ZR. New record of Liriomyza sativae Blanchard (Diptera: Agromyzidae) from China. Entomotaxonomia. 1996;18:311–312. [Google Scholar]
- Wen JZ, Lei ZR, Wang Y. Survey of Liriomyza huidobrensis in Yunnan Province and Guizhou Province, China. Plant Prot. 1998;24:18–20. [Google Scholar]
- Wiederrecht G, Seto D, Parker CS. Isolation of the gene encoding the S. cerevisiae heat shock transcription factor. Cell. 1988;6:841–853. doi: 10.1016/S0092-8674(88)91197-X. [DOI] [PubMed] [Google Scholar]
- Wu C. Heat shock transcription factors: structure and regulation. Annu Rev Cell Dev Biol. 1995;1:441–469. doi: 10.1146/annurev.cb.11.110195.002301. [DOI] [PubMed] [Google Scholar]
- Xiang JC, Lei ZR, Wang HH. Interspecific competition among three invasive Liriomyza species. Acta Ecol Sin. 2012;32:1616–1622. doi: 10.5846/stxb201101140077. [DOI] [Google Scholar]
- Yan H, Zhang S, Li XY, Yuan FH, Qiu W, Chen YG, Weng SP, He JG, Chen YH. Identification and functional characterization of heat shock transcription factor1 in Litopenaeus vannamei. Fish Shellfish Immun. 2014;37:184–192. doi: 10.1016/j.fsi.2014.01.020. [DOI] [PubMed] [Google Scholar]
- Zhang XX, Qin J, Yuan JW, Lu MX, Du YZ. Cloning of a new HSP70 gene from western flower thrips, Frankliniella occidentalis, and expression patterns during thermal stress. Peer J. 2019;7:e7687. doi: 10.7717/peerj.7687. [DOI] [PMC free article] [PubMed] [Google Scholar]