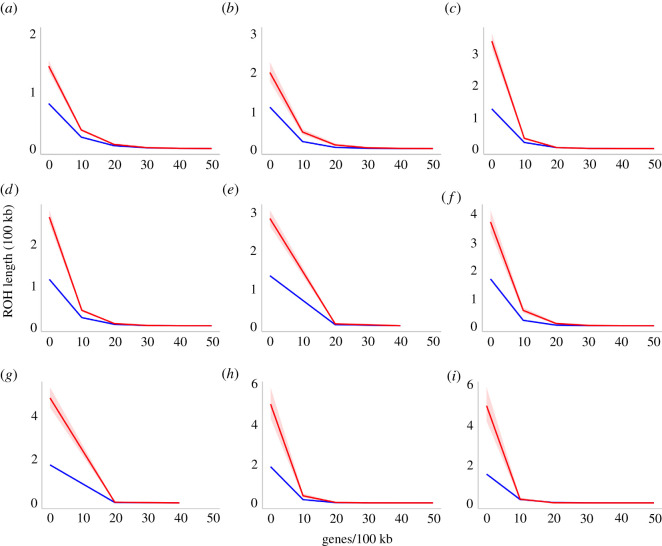
Figure 3.
Predicted values (marginal means) of the lengths of runs of homozygosity (ROH) in hundreds of kilobases (100 kb) that include Ninteract genes (red) are far longer than those that include only other genes (blue) across most biologically relevant gene densities (genes/100 kb). Model results are presented for one or more genome from (a) M. nemestrina from Borneo, (b) M. nemestrina from Sumatra, (c) M. maura, (d) M. tonkeana, (e) M. hecki, (f) M. nigrescens, (g) M. nigra, (h) M. brunnescens and (i) M. togeanus, respectively. Shading indicates the 95% confidence interval of the predicted values. (Online version in colour.)