Abstract
The clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) system was originally discovered in prokaryotes and functions as part of the adaptive immune system. The experimental research of many scholars, as well as scientific and technological advancements, has allowed prokaryote-derived CRISPR/Cas genome-editing systems to transform our ability to manipulate, detect, image, and annotate specific DNA and RNA sequences in the living cells of diverse species. Through modern genetic engineering editing technology and high-throughput gene sequencing, we can edit and splice covalently closed circular DNA to silence it, and correct the mutation and deletion of liver cancer genes to achieve precise in situ repair of defective genes and prohibit viral infection or replication. Such manipulations do not destroy the structure of the entire genome and facilitate the cure of diseases. In this review, we discussed the possibility that CRISPR/Cas could be used as a treatment for patients with liver cancer caused by hepatitis B virus infection, and reviewed the challenges incurred by this effective gene-editing technology.
Keywords: hepatitis B virus DNA, liver cancer, gene editing, CRISPR/Cas9 system
Introduction
Chronic hepatitis B virus (HBV) infection is a common cause of liver disease worldwide. According to the global cancer statistics report released by the World Health Organization in 2018, liver cancer is the fourth leading cause of cancer-related deaths, ranking sixth among all cancers. In 2018, there were ∼841 000 new cases of liver cancer worldwide and 781 000 deaths, with 393 000 new cases and 369 000 deaths each year in China. HBV infection is a progressive infectious disease, and the condition can gradually evolve into liver failure, cirrhosis, and even liver cancer, which seriously endangers the physical and mental health of patients. Using clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) gene-editing technology, scientists designed a specific single-stranded guide RNA (sgRNA) targeting covalently closed circular DNA (cccDNA). The double-stranded structure of the cccDNA is cut by the Cas9, which damages the cccDNA activity, resulting in error repair1 and leading to an inability to transcribe the required mRNA, which may even cease being active, in an effort to cure HBV infection.2 In addition, we can screen out overexpressed genes in liver cancer through gene sequencing and use the CRISPR/Cas9 system to perform gene editing and silence them. It is obvious that utilizing CRISPR/Cas9 to construct liver cancer disease models and look for potential diagnostic and therapeutic targets is worth exploring.3 Using this technique, we can propose new ideas for clinical treatment, create individualized treatments for patients with liver cirrhosis or liver cancer caused by hepatitis B infection, and improve patient prognosis and healing.4,5 Cong et al6 designed 2 different type II CRISPR/Cas systems and demonstrated that Cas9 nuclease can be guided by short RNA to induce precise cleavage at endogenous genomic sites in human and mouse cells. Cas9 can also be converted into a gap enzyme, which promotes homology-oriented repair with minimal mutagenic activity. Finally, multiple guide sequences can be encoded into a single CRISPR array, which enables the simultaneous editing of several sites in the mammalian genome, thereby demonstrating the easy programmability and wide applicability of RNA-guided nuclease technology.6,7
Phenotype and Occurrence of Liver Cancer
Primary liver cancer refers to malignant tumors originating from liver cells or intrahepatic bile duct epithelial cells and includes 3 different histopathological types: hepatocellular carcinoma (HCC), intrahepatic cholangiocarcinoma, and mixed type.
In Asia, HBV infection is the main cause of liver cancer, whereas, in Europe and America, hepatitis C virus (HCV) infection is the causative factor of this disease. During infection, the DNA sequence of HBV and that of the host cell are destroyed or reintegrated so that the oncogene and tumor suppressor gene are activated and inactivated, respectively, resulting in the cancerous transformation of the cell.8 The carcinogenic mechanism of hepatitis C is related to HCV sequence variation. HCV continuously infects liver cells through sequence variation to avoid immune recognition, causing long-term inflammation of the liver, recurring liver cell necrosis, and regeneration. Subsequently, chronic liver infection promotes the occurrence of liver fibrosis due to the wound healing response to tissue damage, followed by the accumulation of gene mutations, destruction of the dynamic balance of cell proliferation, leading to cell cancelation.9 Simultaneously, aflatoxins, liver fibrosis, long-term exposure to nitrous acid, vinyl chloride, and other factors are also important risk factors for liver cancer.
Mechanism of the CRISPR/Cas9 System
The CRISPR/Cas9 system is a prokaryotic immune system. It is a family of extraordinary DNA repeats that are widely found in the genomes of bacteria and archaea and facilitate resistance against the invasion of foreign genetic materials, such as phage viruses and foreign plasmids.10,11 This is similar to the principle of RNA interference (RNAi) in eukaryotes. It is precisely because of this function that the CRISPR/Cas9 system has been developed as an efficient gene-editing tool.12 In 2008, mature CRISPR RNAs (crRNAs) were shown to serve as guides in a complex with Cas proteins to interfere with virus proliferation in Escherichia coli.13 The same year, the DNA targeting activity of the CRISPR/Cas system was reported in the pathogen Staphylococcus epidermidis.14
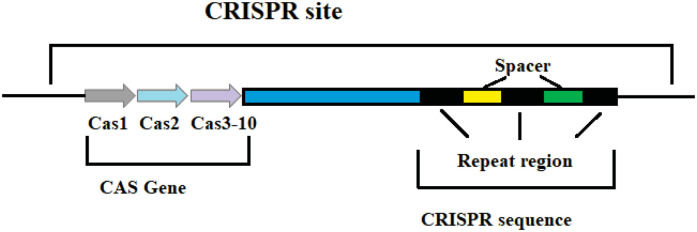
The CRISPR sequence is composed of many short and conserved repeat regions and spacer regions. The repeat sequence region contains palindrome sequences that can form a hairpin structure. The spacer sequence of the CRISPR/Cas9 system is very special. They are foreign DNA sequences captured by bacteria.15,16 There is a polymorphic family gene upstream, and the protein encoded by this gene can interact with the CRISPR sequence region. Therefore, this gene was named CRISPR-associated gene (CRISPR associated, Cas)17,18 (Figure 1).
Figure 1.
Structure of the CRISPR/Cas9 system.
Abbreviations: CRISPR, clustered regularly interspaced short palindromic repeats; Cas9, CRISPR-associated protein 9.
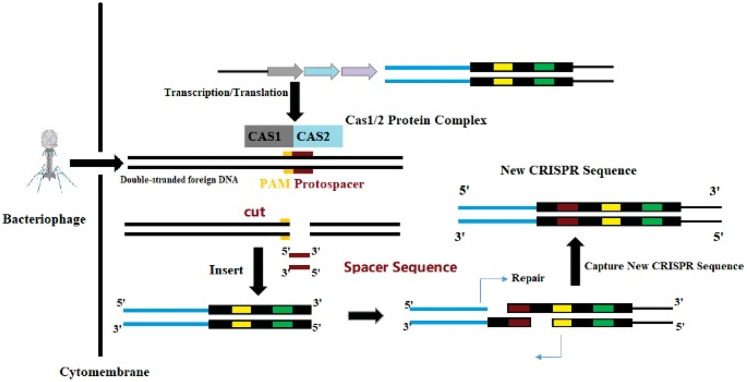
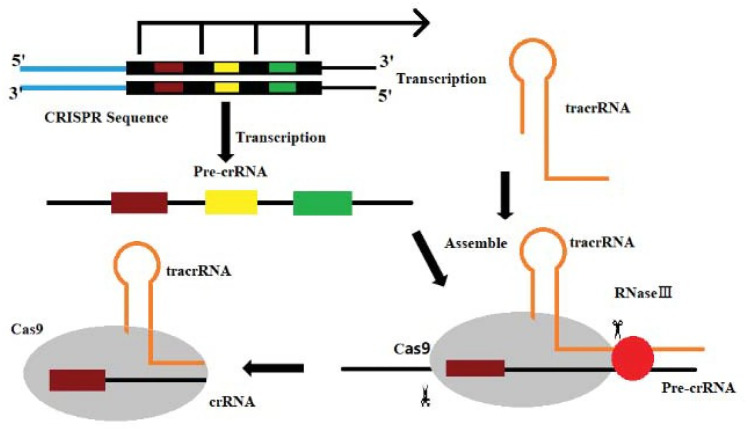
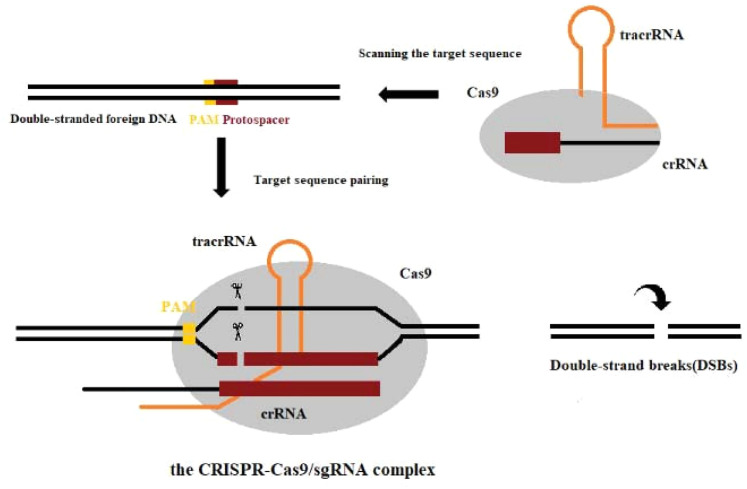
Among the different types of CRISPR/Cas systems, type II CRISPR/Cas9 is the most widely used and studied system.19 Several bases extending to both ends of the original spacer are very conserved and are called protospacer adjacent motifs (PAMs). A PAM usually consists of 3 NGG bases (N is any base). Some researchers have found that PAM has an allosteric effect, triggering the interdependent conformational dynamics of the CaS9 catalytic domains (HNH and RuvC) responsible for the cooperative cutting of two DNA strands.20 When the virus invades, the Cas1/2 protein complex cuts the original spacer sequence from the foreign DNA and inserts the original spacer sequence downstream of the adjacent CRISPR sequence with the assistance of other enzymes.21 Thus, a new spacer sequence is added to the CRISPR sequence of the genome. New spacers then provide sequence-specific memory against their corresponding invading plasmids or viruses (Figure 2).22,23 The CRISPR sequence will transcribe 2 products under the control of the leader region: pre-CRISPR-derived RNA and trans-acting crRNA (tracrRNA). Then, this is further processed into mature crRNA and together with tracrRNA and the proteins encoded by Cas9, complexes are assembled (Figure 3).13 Finally, crRNAs act as guides to specifically target the PAM, while Cas9 cuts the matched DNA. In the type II CRISPR/Cas9 system, the sgRNA–Cas9 complex binds to its target DNA to ensure that Cas9 cuts the two strands of DNA, thereby preventing the spread of foreign DNA (Figure 4).24,25 The CRISPR/Cas system is a powerful tool for accurate gene editing. In addition to basic editing methods such as gene knockout and gene replacement, it can also be used for gene activation, disease model construction, and even gene therapy.26
Figure 2.
The CRISPR/Cas9 system used to obtain foreign DNA.
Abbreviations: CRISPR, clustered regularly interspaced short palindromic repeats; Cas9, CRISPR-associated protein 9.
Figure 3.
Cas9–tracrRNA–crRNA complex.
Abbreviations: Cas9, CRISPR-associated protein 9; tracrRNA, trans-acting crRNA; crRNA, CRISPR RNA.
Figure 4.
The CRISPR/Cas9 system cutting foreign DNA.
Abbreviations: CRISPR, clustered regularly interspaced short palindromic repeats; Cas9, CRISPR-associated protein 9.
In recent studies, some scholars have discovered that CRISPR-Cas < D from macrophages is an ultracompact genome editor, which has a stronger target recognition ability than other CRISPR/Cas proteins. Cas < D is useful for genome editing and DNA detection. However, its molecular weight is half that of Cas9 and Cas12a genome-editing enzymes, which gives Cas < D an advantage in cell delivery, thus expanding the genome-editing toolbox.27
Delivery of the CRISPR/Cas9 System in Liver Cells
At present, the delivery of the CRISPR/Cas9 system can be divided into 3 levels: plasmid, RNA, and protein delivery.28-30
Among them, the delivery of plasmids encoding Cas9 protein and sgRNA is at the DNA level,31 which is convenient, fast, and has high stability. However, the delivery of plasmids reduces the efficiency of gene editing, prolongs the cleavage time of Cas9 protein, and increases the off-target rate and the risk of gene integration. To improve the gene-editing efficiency of plasmid transfection, Ghassemi32 constructed a plasmid encoding Cas9 protein and double sgRNA, delivered it to donor-fertilized eggs by microinjection, and implanted the fertilized eggs into pregnant mice for offspring screening and genotyping. Finally, a β-thalassemia mouse model with Hobbs-bs gene knockout was established.32
At the RNA level, the delivery of mRNA encoding Cas9 protein has a lower off-target rate than a delivery plasmid. It only needs to be delivered to the liver cytoplasm and then translated into Cas9 protein under the action of ribosomes in the liver cytoplasm. The limitation of this delivery method is that the mRNA itself is poorly stable and susceptible to degradation by RNase both in vivo and in vitro. Therefore, a suitable delivery method is required to protect mRNA from degradation by enzymes. Research has shown that new delivery technology is reliable and effective. It has been reported that zwitterionic amino lipids (ZALs) are uniquely able to (co)deliver long RNAs, including Cas9 mRNA and sgRNAs. Under the editing of the CRISPR/Cas9 system, the ZAL nanoparticle (ZNP) delivery of low sgRNA doses (15 nm) reduces protein expression by >90% in cells. In contrast to transient therapies (such as RNAi), research has shown that the ZNP delivery of sgRNA enables permanent DNA editing with an indefinitely sustained 95% decrease in protein expression.33
The direct delivery of the Cas9 protein at the protein level has certain advantages over DNA or RNA delivery methods. Its immunogenicity is low and there is no potential problem of permanently integrating CRISPR genes into the human genome, with low off-target rates and low toxicity. However, the encapsulation size of sgRNA and Cas9 protein is too large, which makes it difficult to introduce into hepatocytes.34 In terms of viral vectors, common viral vectors in the CRISPR/Cas9 system include recombinant adeno-associated virus(AAV) vectors,35 bacteriophages, and adenoviruses. Although viral vectors can effectively deliver the CRISPR/Cas9 system through cell transduction, they may still cause unnecessary immunogenicity and mutation risks in humans, thereby limiting their clinical transformation.36 In contrast, research has shown that in recent years, nonviral vectors have effectively achieved delivery to cells and tissues in vivo and in vitro; liposomes,36 nanocarriers,33,37 cell-penetrating peptides,38 etc, have gradually become potential tools for CRISPR/Cas9 system gene therapy.39,40
Therefore, any nonviral vector should have good tolerance, biocompatibility, and nonimmunogenicity, and should be able to deliver effective Cas9–sgRNA to the nucleus for genome editing.40 So far, the relatively low gene delivery efficiency and transgene expression have been the major barriers in nonviral vector-based gene therapy. However, with the rapid development of novel biomaterials in recent years, the efficient delivery of gene payloads to pass multiple barriers under physiological conditions and promote transgene expression can be achieved.41
Emerging delivery strategies also make the delivery of the CRISPR/Cas9 system more mature. Precise control over the CRISPR/Cas9 system in temporal and spatial resolution is essential for gene regulation and editing.42 Here, we presented a novel light-controlled crRNA via coupling vitamin E and a photolabile linker at the 5′-terminus to inactivate the CRISPR/Cas9 system.43 Vitamin E modification did not affect the ribonucleoprotein (RNP) formation of the Cas9/crRNA/tracrRNA complexes, but did inhibit the association of RNP with the target DNA. Upon light irradiation, vitamin E-caged crRNA was successfully activated to achieve the light-induced genome editing of vascular endothelial cell growth factor A in human cells through T7E1 assay and Sanger sequencing as well as the gene knockdown of enhanced green fluorescent protein (EGFP) expression in EGFP stably expressing cells. This new caging strategy for crRNA is readily achievable. This may provide new methods for the spatiotemporal photo regulation of CRISPR/Cas9-mediated gene editing, which reduces uncontrollable risks encountered in the delivery and editing of CRISPR/Cas9.43,44
The Mechanism of the CRISPR/Cas9 System in cccDNA
HBV is an enveloped DNA virus that explicitly targets human liver cells and replicates by the reverse transcription of a terminally redundant viral RNA, the progenome. Upon infection, the circular, partially double-stranded virion DNA is converted in the nucleus to a covalently closed circular DNA (cccDNA) that assembles into a minichromosome, the template for viral mRNA synthesis.45 HBV infects liver cells through the interaction between viral envelope protein determinants and heparan sulfate proteoglycan46 on the liver cell surface. This interaction brings it close to the sodium ion taurocholic acid cotransport peptide (NTCP)47,48; the high-affinity action that produces the bile acid-binding pocket of NTCP promotes the entry of the virus. The complete HBV life cycle includes attachment, entry, uncoating, trafficking to the nucleus, covalently closed circular DNA (cccDNA) formation, transcription, translation, encapsulation, assembly, and secretion. Significant HBV infection requires the virus to penetrate the cell membrane and then unshell in the cytoplasm. Its nucleic acid components migrate in the endoplasmic reticulum structure of the cell.49 Part of the nucleic acid components eventually enters the cell nucleus, where HBV DNA can also interact with the liver cell nucleus. The chromosomal DNA becomes integrated into HBV DNA. Of course, a part of HBV DNA is in a free state to prepare for the virus replication process. Within a few hours of HBV-infected cells, the virus is uncoated in the cytoplasm, showing a relaxed state, and the incomplete double-stranded HBV DNA enters the nucleus. Under the catalysis of HBV DNA polymerase, the HBV DNA gap is filled with a single nucleotide, thereby forming a complete double-stranded circular structure, then forming an HBV DNA helical shape, and then folding into supercoiled DNA, cccDNA, forming the HBV DNA group replication original template.50,51
HBV cccDNA is stable in infected cells and has a long half-life.51 Current knowledge about the formation and degradation of cccDNA is still limited. In addition, HBV infection has further oncogenic potential after HBV integration into the host genome, which is an additional pathway contributing to HCC. HBV infection is managed with reverse transcriptase inhibitors, which are nucleoside or nucleotide analogs, and, less commonly, with interferon therapy.52,53 The total elimination of HBV from patients (HBV cure) remains a distant treatment goal, but a more practical goal is the achievement of hepatitis B surface antigen (HBsAg) serum clearance (functional cure) as early as possible after infection.53,54 As a marker of a complete cure, its removal has been challenging to achieve in traditional antiviral methods. At present, genetic engineering, as an emerging treatment method, is expected to overcome this problem.55-57 The CRISPR/Cas9 system is mainly composed of Cas9 protein and single-stranded guide sgRNA. The Cas9 protein has the function of cutting double-stranded DNA, while sgRNA plays a guiding role. In the presence of the adjacent PAM, the Cas9 protein can reach different target sites through complementary base pairing under the guidance of sgRNA and cut the target gene to achieve DNA double-strand breaks (DSBs).58-60 Some researchers initially evaluated the HBV genome target and potential targets, used the whole genome sequence of HBV genotype obtained from GenBank, and then designed sgRNA for the target gene region, together with the DNA encoding the Cas9 protein. Currently, a large number of sgRNAs have been developed and reported based on their high preservation among HBV genotypes and significant effects have been found.61 The fragments are integrated into the delivery plasmid and transfected into HBV-infected liver cells at the same time. The relative content of HBV pregenome RNA and HBsAg (S-RNA) mRNA is determined by polymerase chain reaction analysis, and the relative changes in the total HBV DNA concentration and cccDNA level in the cell are obtained. Under the action of the CRISPR/Cas9 system, the broken DNA double-strands undergo error repair or artificially induced gene mutations, so that cccDNA loses its biological activity or even degrades and its cell content is reduced, thereby achieving the purpose of curing hepatitis B.62,63 The results of a recent study have been encouraging, as they showed that humanized liver mouse models have been developed that accurately reflect human HBV infection. These models use immunodeficient mice with a genetic background lethal for mouse hepatocytes; transplanted human hepatocytes are provided to reconstitute the liver, which becomes chimeric. Researchers have harnessed AAV vectors and S aureus (Sa) CRISPR/Cas9 to edit the HBV genome in liver-humanized FRG mice chronically infected with HBV and receiving entecavir. HBV-specific AAV-Sa Cas9 therapy significantly improves the survival of human hepatocytes, shows a trend toward decreasing the total liver HBV DNA and cccDNA, and is well tolerated.64 These results are hopeful in terms of curing chronic HBV infection.
Application of the CRISPR/Cas9 System in Tumor Genes of Liver Cancer Caused by HBV
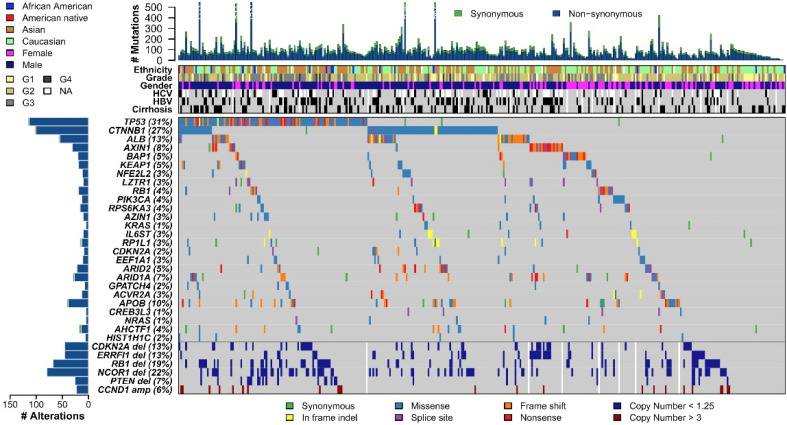
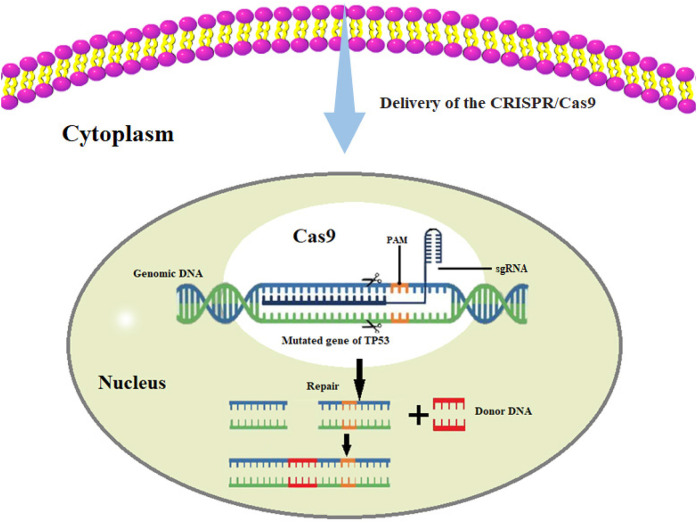
Research on liver cancer caused by HBV infection mainly focuses on the mechanism by which HBV DNA causes HCC. Moreover, the original description of HBV DNA integrated into the host cell genome is the primary HCC tissue and HCC-derived cell lines, which suggests that the integrated HBV DNA is related to tumorigenesis.65,66 The reported mechanisms include (1) the cis-mediated insertional mutagenesis of HCC-associated genes, (2) the induction of chromosomal instability by integrated DNA, and (3) the expression of mutant HBV genes from the persistent integrated form.67 However, the mechanism of HBV-induced HCC carcinogenesis remains unclear and poorly characterized.68 Nowadays, we can use high-throughput sequencing to obtain single-base and multibase variants in the DNA sequence of liver cancer tissues, copy number variations, genome structure variations, etc,68-70 and then pass CRISPR/Cas9 system-mediated homologous recombination, knocking a therapeutic transgene at the desired location to induce cancer cell death, or induce targeted double-stranded DNA breaks in the genome through the CRISPR/Cas9 system to achieve genetic modification.71 Wheeler et al investigated 363 HCC cases using whole-exome sequencing and DNA copy number analyses and 196 HCC cases through DNA methylation, RNA, miRNA, and proteomic expression analyses. DNA sequencing and mutation analysis identified significantly mutated genes, including TP53, LZTR1, and EEF1A1 (Figure 5). Reprinted from Cancer Genome Atlas Research Network. Electronic address: wheeler@bcm.edu; cancer genome atlas research network,72 with permission from Elsevier. It is well established that TP53 is mutated in many cancers, and liver cancer is no exception. When the DNA of normal liver cells is damaged, the P53 protein prevents the cell cycle from arresting at the G1/S phase and repairs the damage. If it cannot be repaired, P53 promotes cell apoptosis. In TP53-mutated liver cells, the cell cycle is uncontrolled, and cells continue to divide even under adverse conditions, eventually leading to the occurrence of liver cancer. If one can design a sgRNA that recognizes PAM, this would allow the Cas9 protein to successfully recognize or attach to and repair the mutated base in the TP53 gene, eventually being possible to curb the progression of liver cancer. Through gene-editing tools, we can restore the original functions of TP53, which might prevent liver cancer progression (Figure 6). Furthermore, we can import genes that do not exist in the cells into cells with abnormal functions, such as liver cancer cells, and use the expression products of these genes to treat diseases. Besides, introducing suicide genes to induce cell death by suicide is also an important tumor-treating strategy, in an effort to eliminate tumor cells.73 We can perform genome-wide screening using CRISPR-mediated genome editing to identify inhibitors or critical drivers such as the lncRNA74 of liver tumor formation, and then develop strategies to suppress or reduce related factors to treat patients with liver cancer.73,75,76
Figure 5.
The genomic landscape of liver hepatocellular carcinoma and mutational signatures. Top panel shows individual tumor mutation rates, while the middle panel details ethnicity, tumor grade, age, gender, HCV, and HBV infection status, and cirrhosis for 363 patients with HCC. Bottom panel shows genes with significantly different levels of mutation (MutSig suite, FDR, <0.1), and mutation types are indicated in the legend, at the bottom. The bottom six rows display significant DNA copy number alterations in likely cancer driver genes.
Abbreviations: HCV, hepatitis C virus; HBV, hepatitis B virus; HCC, hepatocellular carcinoma.
Figure 6.
Application of the gene-editing technology in liver cancer.
Recently, some Cas9 family members have been found to have the ability to target RNA as well as DNA. Various RNA-targeting systems for Cas9 (RNA-targeted Cas9 [RCas9]) have been established and have opened up new applications. RCas9 could allow the identification and manipulation of RNA substrates in live cells, thereby empowering the study of cellular gene expression, and could ultimately spawn patient- and disease-specific diagnostic and therapeutic tools,77 such as gene expression silencing at the posttranscriptional level, intracellular transcription interference, and anti-RNA virus infection.78 Researchers recently established the ability of a repurposed type II CRISPR system to target and track RNA in human cells, thereby indicating the potential of other RNA manipulations using RCas9. In a study, researchers describe the ability of CRISPR/Cas9 to both visualize and eliminate MRE RNAs in human cells. The researchers observed the efficient elimination of all MRE RNAs and described a reduced version of the RNA-targeting CRISPR/Cas9 system that was optimized for AAV packaging. They also observed a reversal of disease-associated molecular defects, including the reduction of polyglutamine proteins and the near-complete reversal of DM1-associated splicing defects to resemble healthy patient cells (93% reversal in patient myotubes).79 Thus, we can use the RCas9 system to target and edit abnormally transcribed mRNA in liver cancer tissues, eliminate the defects of related molecular mechanisms in these cells, and even repair them. This will provide more strategies for the treatment of HCC.
In addition, in the current treatment of various tumors and cancers, T-cell receptor (TCR)-engineered T-cells immunity combined with multiple gene-editing technologies will become another emerging method in the medical field.80 This method extracts immune T-cells from patients and then uses genetic engineering technology to write in vitro gene fragments that allow T-cells to recognize and kill specific tumor cells. These TCR-T-cells are cultured in large numbers in the laboratory. Then, the expanded enhanced immune T-cells are returned to the patient for treatment. These edited T-cells are extremely efficient and are associated with an extremely low risk of rejection, being an innovative treatment method.81,82 At present, researchers have reported a first human phase I clinical trial to test the safety and feasibility of T-cells edited by multiple CRISPR/Cas9 systems in 3 patients with refractory cancer. There was evidence of tumor regression in 1 of the patients with the highest cell implantation rate. We believe that this method has broad application potential in the treatment of liver cancer.81,83 Moreover, in the present research, the use of the CRISPR/Cas9 system to simultaneously disrupt multiple genomic loci, resulted in the generation of chimeric antibody receptor engineered T-cell (CART) that were deficient in the expression of endogenous TCR and HLA class I, which can be used as gene-disrupted allogeneic CART and further developed as universal CART cells. Researchers also generated gene-disrupted allogeneic CART cells by combining the lentiviral delivery of CAR and crRNA electroporation to disrupt endogenous TCR and B2M genes simultaneously. In addition, we demonstrated that the disruption of endogenous PD1 enhances the efficacy of gene-disrupted allogeneic CAR therapy in tumor models. The therapeutic value of blocking the PD1 inhibitory pathway was confirmed in these systems by the ablation PD1 of CART cells in a solid tumor model and by the triple ablation of TCR, B2M, and PD1 of CART cells in a leukemia tumor model.83 We believe that TCR-T-cell immunity combined with multiple gene-editing technologies has excellent application value for liver cancer.84 In addition, it can significantly reduce the rejection of T-cells in HCC by the body.
Discussion
At present, the CRISPR/Cas9 system and various other biological technologies are more and more closely connected and developed, which has brought about a new era in the treatment of various difficult diseases.85 However, new technologies are accompanied by new risks. For example, the CRISPR/Cas9 system delivery technology cannot be effectively delivered to the body. The effectiveness and specificity of the delivery method include whether it can specifically act on the target site and the safety of the foreign vector. Immunogenicity measures whether there is a risk of cancer caused by insertion into the genome and whether it will incur an immune response. The editing efficiency of the editing tool itself and the potential off-target effects assess whether the editing tool can effectively edit the corresponding pathogenic site, and the sequence of similar sites, or even other sequences due to the overexpression of action elements.33 Although this type of technology is developing rapidly, it is still in the clinical trial phase, owing to various safety issues. Therefore, improving the safety and feasibility of the CRISPR/Cas9 system should be the future research focus.86 In a recent study, CRISPR/Cas-mediated base editors (BEs) in inactivating HBV gene expression without DNA cleavage. Streptococcus pyogenes Cas9 (SpCas9)-BE with certain gRNAs effectively base-edited polymerase and surface genes and reduced HBV gene expression in cells harboring integrated HBV genomes, but induced very few insertions or deletions (indels).87
Some scholars have proposed a self-restricted CRISPR/Cas9 system, which inserts target gene sequences corresponding to gRNA at both ends of the Cas9 promoter. When a DSB occurs in the target gene sequence, the Cas9 promoter is cut off and transcription stops. The Cas9 protein expression level of the system 60 h after transfection is only 10% of that of the wild-type system, and the frequency of the off-target formation decreases by 76.7%. At the same time, this system does not include additional gRNA sequences, so the possibility of introducing new off-target mutations is reduced. More importantly, this system consists of a single plasmid, which may be easily introduced into the body through viral vectors or nanoparticles. In general, the CRISPR/Cas9 system has broad application potential in the clinical treatment of tumors, but the nontarget effects are the main obstacles that limit the clinical application of the system. Among them, nontarget effects caused by the overexpression of the Cas9 protein will be the most difficult to overcome in research.88 In a study, scientists conducted a high-throughput evaluation of the activity of CRISPR/Cas9 to examine which small-molecule drugs can effectively inhibit its activity. To this end, they built 2 systems. The first system can monitor whether SpCas9 protein can automatically evaluate the changes in fluorescence (indicating that SpCas9 has completed the shearing of DNA); that is, if SpCas9 cannot smoothly bind to DNA under the action of a certain small molecule, or cannot complete the shearing of DNA. This means that this small molecule has effectively inhibited the CRISPR/Cas9 system. Following this idea, the researchers successfully screened out 2 compounds, which can inhibit SpCas9 activity in a dose-dependent manner in mammalian cell lines. However, the researchers also pointed out that these potential compounds cannot be directly used in humans. Next, the researchers will focus on 2 research directions. The first is the determination of the binding site of this type of inhibitor on the SpCas9:gRNA complex, and the examination of its mechanism of action to optimize its effectiveness. The second is to determine whether these molecular inhibitors can act on other targets in mammalian cells and to evaluate their specificity to other CRISPR-related nucleases; in future studies, it is also necessary to confirm whether these molecules will cause other side effects.89
Significance and Future Directions of CRISPR/Cas9
In the technical field, CRISPR/Cas9 has very obvious advantages: the vector construction of the system is simple, and the targeting efficiency is high. The system only requires the construction of a CRISPR sgRNA of dozens of bases to match the DNA sequence, thereby mediating the Cas9 protein to cut the DNA sequence. Moreover, the frequency of CRISPR/Cas9 editable sites is high, and it is easy to choose suitable sites for gene editing; noteworthy, CRISPR/Cas9 can edit the genome at multiple sites simultaneously. The CRISPR/Cas9 technology corrects defective genes by introducing new gene codes, thereby being able to repair certain genetic diseases and improving the success of cancer treatment. Furthermore, this technology can be used to discover and manipulate specific genes and as a daily tool to explore the physiology and diagnosis of diseases. For example, CRISPR/Cas9 can be used to arm human immune cells to fight cancer or HIV infection, repair defective genes in human embryos, and prevent infants from inheriting serious diseases. Thus, CRISPR/Cas9 has significantly changed basic science, produced many impactful results, and will surely allow the development of new treatment methods.
Societal development dictates that research focuses on new materials such as nano- and small-molecule polymers as well as viral vector analogs. Only in this manner can the potential off-target effects of the CRISPR/Cas9 system be reduced as soon as possible. In addition, this technology can allow us to change the spatial conformation of the Cas9 protein through light induction or other means to achieve a controllable enhancement or reduction of gene editing capabilities at any time. It is also important to look for or synthesize small-molecule Cas9 protein inhibitors, to meet the demand for the precise control of Cas9 dosage and time. In addition, by constructing a new inducible Cas9 protein, its expression in cells can be regulated. The aforementioned methods can help reduce the ability of the Cas9 protein to bind nonspecifically to DNA and enhance the competitiveness of the target sequence to bind the Cas9 protein. The efforts of researchers to promote the development of Cas9 in medicine are highly anticipated.
Acknowledgment
This work was supported by the corresponding author and co-authors. At the same time, sincerely thank for their guidance and help on this article.
Abbreviations
- AAV
adeno-associated virus
- BE
base editor
- HBsAg
hepatitis B surface antigen
- CART
chimeric antibody receptor engineered T-cell
- Cas9
CRISPR-associated protein 9
- cccDNA
covalently closed circular DNA
- CRISPR
clustered regularly interspaced short palindromic repeats
- crRNA
CRISPR RNA
- DSB
double-strand break
- EGFP
enhanced green fluorescent protein
- HBV
hepatitis B virus
- HCC
hepatocellular carcinoma
- HCV
hepatitis C virus
- LV
lentiviral
- PAM
protospacer adjacent motifs
- PCR
polymerase chain reaction
- pre-crRNA
pre-CRISPR-derived RNA
- RCas9
RNA-targeted Cas9
- RNAi
RNA interference
- RNP
ribonucleoprotein
- Sa
S aureus
- sgRNA
single-stranded guide RNA
- SpCas9
Streptococcus pyogenes Cas9
- TCR
T-cell receptor
- tracrRNA
trans-acting crRNA
- VEGFA
vascular endothelial cell growth factor A
- ZAL
zwitterionic amino lipid
- ZNP
ZAL nanoparticle.
Footnotes
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by the Natural Science Foundation of China (grant no. 30801088) and scientific research funds for hosting the project “miRNA-146a inhibits HCC by targeting cyclooxygenase-2 and FLAP and its mechanism” as academic and technical leaders and reserve candidates of Anhui Province in 2019 (2019H214).
Ethical Approval: Our study did not require an ethical board approval because it did not contain human or animal trials.
ORCID iD: Tao Li https://orcid.org/0000-0003-3822-1856
References
- 1.Ohno M, Otsuka M, Mishawaka T, Yoshikawa T, Takata A, Koike K. Novel therapeutic approaches for hepatitis B virus covalently closed circular DNA. World J Gastroenterol. 2015;21(23):7084-7088. doi: 10.3748/wig.v21.i23.7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee C. CRISPR/Cas9-based antiviral strategy: current status and the potential challenge. Molecules. 2019;24(7):1349. doi: 10.3390/molecules24071349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alves-Bezerra M, Furey N, Johnson CG, Bissig KD. Using CRISPR/Cas9 to model human liver disease. JHEP Rep. 2019;1(5):392-402. doi: 10.1016/j.jhepr.2019.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen S, Sun H, Miao K, Deng CX. CRISPR-Cas9: from genome editing to cancer research. Int J Biol Sci. 2016;12(12):1427-1436. doi: 10.7150/ijbs.17421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Torres-Ruiz R, Rodriguez-Perales S. CRISPR-Cas9 technology: applications and human disease modelling. Brief Funct Genomics. 2017;16(1):4-12. doi: 10.1093/bfgp/elw025. [DOI] [PubMed] [Google Scholar]
- 6.Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas. Systems. Science. 2013;339(6121):819-823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mali P, Yang L, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823-826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levrero M, Zucman-Rossi J. Mechanisms of HBV-induced hepatocellular carcinoma. J Hepatol. 2016;64(1 Suppl): S84-S101. doi: 10.1016/j.jhep.2016.02.021. [DOI] [PubMed] [Google Scholar]
- 9.Khatun M, Ray RB. Mechanisms underlying hepatitis C virus-associated hepatic fibrosis. Cells. 2019;8(10):1249. doi: 10.3390/cells8101249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346(6213):1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 11.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816-821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pickar-Oliver A, Gersbach CA. The next generation of CRISPR-Cas technologies and applications. Nat Rev Mol Cell Biol. 2019;20(8):490-507. doi: 10.1038/s41580-019-0131-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brouns SJ, Jore MM, Lundgren M, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321(5891):960–964. 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322(5909):1843-1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482(7385):331-338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 16.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32(4):347-355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327(5962):167-170. doi: 10.1126/science.1179555. PMID: 20056882. [DOI] [PubMed] [Google Scholar]
- 18.Jiang F, Doudna JA. CRISPR-Cas9 structures and mechanisms. Annu Rev Biophys. 2017;46:505-529. doi: 10.1146/annurev-biophys-062215-010822. [DOI] [PubMed] [Google Scholar]
- 19.Makarova KS, Wolf YI, Alkhnbashi OS, et al. An updated evolutionary classification of CRISPR–Cas systems. Nat Rev Microbiol. 2015;13(11):722-736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palermo G, Ricci CG, Fernando A, et al. Protospacer adjacent motif-induced allostery activates CRISPR-Cas9. J Am Chem Soc. 2017;139(45):16028-16031. doi: 10.1021/jacs.7b05313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grainy J, Garrett S, Graveley BR, P Terns M. CRISPR repeat sequences and relative spacing specify DNA integration by Pyrococcus furiosus Cas1 and Cas2. Nucleic Acids Res. 2019;47(14):7518-7531. doi: 10.1093/nar/gkz548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157(6):1262-1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yosef I, Goren MG, Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012;40(12):5569-5576. doi: 10.1093/nar/gks216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Westra ER, Buckling A, Fineran PC. CRISPR-Cas systems: beyond adaptive immunity. Nat Rev Microbiol. 2014;12(5):317-326. doi: 10.1038/nrmicro3241. [DOI] [PubMed] [Google Scholar]
- 25.Liu C, Zhang L, Liu H, Cheng K. Delivery strategies of the CRISPR-Cas9 gene-editing system for therapeutic applications. J Control Release. 2017;266:17-26. doi: 10.1016/j.jconrel.2017.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343(6166):80-84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pausch P, Al-Shayeb B, Bisom-Rapp E, et al. CRISPR-Cas<D from huge phages is a hypercompact genome editor. Science. 2020;369(6501):333-337. doi: 10.1126/science.abb1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eoh J, Gu L. Biomaterials as vectors for the delivery of CRISPR–Cas9. Biomater Sci. 2019;7(4):1240–1261. 10.1039/c8bm01310a [DOI] [PubMed] [Google Scholar]
- 29.Luther DC, Lee YW, Nagaraj H, Scaletti F, Rotello VM. Delivery approaches for CRISPR/Cas9 therapeutics in vivo: advances and challenges. Expert Opin Drug Deliv. 2018;15(9):905-913. doi: 10.1080/17425247.2018.1517746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li L, Hu S, Chen X. Non-viral delivery systems for CRISPR/Cas9-based genome editing: challenges and opportunities. Biomaterials. 2018;171:207-218. doi: 10.1016/j.biomaterials.2018.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rui Y, Varanasi M, Mendes S, Yamagata HM, Wilson DR, Green JJ. Poly (beta-amino ester) nanoparticles enable nonviral delivery of CRISPR-Cas9 plasmids for gene knockout and gene deletion. Mol Ther Nucleic Acids. 2020;20:661-672. doi: 10.1016/j.omtn.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghassemi B, Shamsara M, Soleimani M, Kiani J, Rassoulzadegan M. Pipeline for the generation of gene knockout mice using dual sgRNA CRISPR/Cas9-mediated gene editing. Anal Biochem. 2019;568:31-40. doi: 10.1016/j.ab.2018.12.002. Erratum in: Anal Biochem. 2019 Oct 15. [DOI] [PubMed] [Google Scholar]
- 33.Miller JB, Zhang S, Kos P, et al. Non-viral CRISPR/Cas gene editing in vitro and in vivo enabled by synthetic nanoparticle Co-delivery of Cas9 mRNA and sgRNA. Angew Chem, Int Ed. 2016;56(4):1059-1063. doi: 10.1002/anie.201610209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zuris JA, Thompson DB, Shu Y, et al. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat Biotechnol. 2014;33(1):73-80. doi: 10.1038/nbt.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun JY, Anand-Jawa V, Chatterjee S, Wong KK. Immune responses to adeno associated virus and its recombinant vectors. Gene Ther. 2003;10(11):964-976. doi: 10.1038/sj.gt.3302039. [DOI] [PubMed] [Google Scholar]
- 36.Liu J, Chang J, Jiang Y, et al. Fast and efficient CRISPR/Cas9 genome editing in vivo enabled by bioreducible lipid and messenger RNA nanoparticles. Adv Mater. 2019;31(33):e1902575. doi: 10.1002/adma.201902575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finn JD, Smith AR, Patel MC, et al. A single administration of CRISPR/Cas9 lipid nanoparticles achieves robust and persistent in vivo genome editing. Cell Rep. 2018;22(9):2227-2235. 10.1016/j.celrep.2018.02.014 [DOI] [PubMed] [Google Scholar]
- 38.Suresh B, Ramakrishna S, Kim H. Cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA for genome editing. Methods Mol Biol. 2017;1507:81-94. doi: 10.1007/978-1-4939-6518-2_7 [DOI] [PubMed] [Google Scholar]
- 39.Glass Z, Lee M, Li Y, Xu Q. Engineering the delivery system for CRISPR-based genome editing. Trends Biotechnol. 2018;36(2):173-185. 10.1016/j.tibtech.2017.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mashel TV, Tarakanchikova YV, Muslimov AR, et al. Overcoming the delivery problem for therapeutic genome editing: current status and perspective of non-viral methods. Biomaterials. 2020;258:120282. doi: 10.1016/j.biomaterials.2020.120282 [DOI] [PubMed] [Google Scholar]
- 41.Li L, He ZY, Wei XW, Gao GP, Wei YQ. Challenges in CRISPR/CAS9 delivery: potential roles of nonviral vectors. Hum Gene Ther. 2015;26(7):452-462. doi: 10.1089/hum.2015.069. [DOI] [PubMed] [Google Scholar]
- 42.Mintzer MA, Simanek EE. Nonviral vectors for gene delivery. Chem Rev. 2009;109(2):259-302. doi: 10.1021/cr800409e. [DOI] [PubMed] [Google Scholar]
- 43.Gangopadhyay SA, Cox KJ, Manna D, et al. Precision control of CRISPR-Cas9 using small molecules and light. Biochemistry. 2019;58(4):234-244. doi: 10.1021/acs.biochem.8b01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, Ling XY, Su XX, et al. Optical control of CRISPR-Cas9 system for gene editing using photolabile crRNA. Angew Chem, Int Ed. 2020;59(47):20895–20899. 10.1002/anie.202009890 [DOI] [PubMed] [Google Scholar]
- 45.Seeger C, Mason WS. Molecular biology of hepatitis B virus infection. Virology 2015;479-480:672-686. doi: 10.1016/j.virol.2015.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schulze A, Gripon P, Urban S. Hepatitis B virus infection initiates with a large surface protein-dependent binding to heparan sulfate proteoglycans. Hepatology. 2007;46(6):1759-1768. doi: 10.1002/hep.21896. [DOI] [PubMed] [Google Scholar]
- 47.Urban S, Bartenschlager R, Kubitz R, Zoulim F. Strategies to inhibit entry of HBV and HDV into hepatocytes. Gastroenterology. 2014;147(1):48-64. doi: 10.1053/j.gastro.2014.04.030. [DOI] [PubMed] [Google Scholar]
- 48.Sun Y, Qi Y, Peng B, Li W. NTCP-reconstituted in vitro HBV infection system. Hepatitis B Virus. 2016;1540:1-14. doi: 10.1007/978-1-4939-6700-1_1 [DOI] [PubMed] [Google Scholar]
- 49.Liang TJ. Hepatitis B: the virus and disease. Hepatology. 2009;49(5 Suppl):S13-S21. doi: 10.1002/hep.22881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nassal M. HBV cccDNA: viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut. 2015;64(12):1972-1984. doi: 10.1136/gutjnl-2015-309809. [DOI] [PubMed] [Google Scholar]
- 51.Bloom K, Maepa MB, Ely A, Arbuthnot P. Gene therapy for chronic HBV—can we eliminate cccDNA? Genes (Basel). 2018;9(4):207. doi: 10.3390/genes9040207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang HC, Kao JH. Persistence of hepatitis B virus covalently closed circular DNA in hepatocytes: molecular mechanisms and clinical significance. Emerg Microbes Infect. 2014;3(9):e64. doi: 10.1038/emi.2014.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xia Y, Liang TJ. Development of direct-acting antiviral and host-targeting agents for treatment of hepatitis B virus infection. Gastroenterology. 2019;156(2):311-324. doi: 10.1053/j.gastro.2018.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trépo C, Chan HL, Lok A. Hepatitis B virus infection. Lancet. 2014;384(9959):2053-2063. doi: 10.1016/S0140-6736(14)60220-8. [DOI] [PubMed] [Google Scholar]
- 55.Seeger C, Sohn JA. Targeting hepatitis B virus with CRISPR/Cas9. Mol Ther Nucleic Acids. 2014;3(12):e216. doi: 10.1038/mtna.2014.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moyo B, Bloom K, Scott T, Ely A, Arbuthnot P. Advances with using CRISPR/Cas mediated gene editing to treat infections with hepatitis B virus and hepatitis C virus. Virus Res. 2018;244:311-320. doi: 10.1016/j.virusres.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 57.Ruiz de Galarreta M, Lujambio A. Therapeutic editing of hepatocyte genome in vivo. J Hepatol. 2017;67(4):818-828. doi: 10.1016/j.jhep.2017.05.012. [DOI] [PubMed] [Google Scholar]
- 58.Liu Y, Zhao M, Gong M, et al. Inhibition of hepatitis B virus replication via HBV DNA cleavage by Cas9 from Staphylococcus aureus. Antiviral Res. 2018;152:58-67. doi: 10.1016/j.antiviral.2018.02.011. [DOI] [PubMed] [Google Scholar]
- 59.Dong C, Qu L, Wang H, Wei L, Dong Y, Xiong S. Targeting hepatitis B virus cccDNA by CRISPR/Cas9 nuclease efficiently inhibits viral replication. Antiviral Res. 2015;118:110-117. doi: 10.1016/j.antiviral.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 60.Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507(7490):62-67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kayesh MEH, Amako Y, Hashem MA, et al. Development of an in vivo delivery system for CRISPR/Cas9-mediated targeting of hepatitis B virus cccDNA. Virus Res. 2020;290:198191. doi: 10.1016/j.virusres.2020.198191. [DOI] [PubMed] [Google Scholar]
- 62.Kostyushev D, Brezgin S, Kostyusheva A, Zarifyan D, Goptar I, Chulanov V. Orthologous CRISPR/Cas9 systems for specific and efficient degradation of covalently closed circular DNA of hepatitis B virus. Cell Mol Life Sci. 2019;76(9):1779-1794. 10.1007/s00018-019-03021-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ramanan V, Shlomai A, Cox DB, et al. CRISPR/Cas9 cleavage of viral DNA efficiently suppresses hepatitis B virus. Sci Rep. 2015;5(10833). doi: 10.1038/srep10833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stone D, Long KR, Loprieno MA, et al. (2020). CRISPR/Cas9 gene editing of hepatitis B virus in chronically infected humanized mice. Molecular Therapy—Methods & Clinical Development. 20:258-275. 10.1016/j.omtm.2020.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Edman JC, Gray P, Valenzuela P, Rall LB, Rutter WJ. Integration of hepatitis B virus sequences and their expression in a human hepatoma cell. Nature. 1980;286(5772):535-538. doi: 10.1038/286535a0. [DOI] [PubMed] [Google Scholar]
- 66.Brechot C, Pourcel C, Louise A, Rain B, Tiollais P. Presence of integrated hepatitis B virus DNA sequences in cellular DNA of human hepatocellular carcinoma. Nature. 1980;286(5772):533-535. doi: 10.1038/286533a0. [DOI] [PubMed] [Google Scholar]
- 67.Tu T, Budzinska MA, Shackel NA, Jilbert AR. Conceptual models for the initiation of hepatitis B virus-associated hepatocellular carcinoma. Liver Int. 2015;35(7):1786-1800. doi: 10.1111/liv.12773. [DOI] [PubMed] [Google Scholar]
- 68.Tu T, Budzinska MA, Shackel NA, Urban S. HBV DNA integration: molecular mechanisms and clinical implications. Viruses. 2017;9(4):75. doi: 10.3390/v9040075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nakagawa H, Fujita M, Fujimoto A. Genome sequencing analysis of liver cancer for precision medicine. Semin Cancer Biol. 2019;55:120-127. doi: 10.1016/j.semcancer.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 70.Craig AJ, von Felden J, Garcia Lezana T, Sarcognato S, Villanueva A. Tumour evolution in hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2020;17(3):139-152. doi: 10.1038/s41575-019-0229-4. [DOI] [PubMed] [Google Scholar]
- 71.Wang H, La Russa M, Qi LS. CRISPR/Cas9 in genome editing and beyond. Annu Rev Biochem. 2016;85:227-264. doi: 10.1146/annurev-biochem-060815-014607. [DOI] [PubMed] [Google Scholar]
- 72.Wheeler DA, Roberts LR, Sheth Margi, et al. Electronic address: wheeler@bcm.edu; cancer genome atlas research network. Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell. 2017;169(7):13271341. e23. doi: 10.1016/j.cell.2017.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Khan N, Bammidi S, Chattopadhyay S, Jayandharan GR. Combination suicide gene delivery with an adeno-associated virus vector encoding inducible caspase-9 and a chemical inducer of dimerization Is effective in a Xenotransplantation model of hepatocellular carcinoma. Bioconjug Chem. 2019;30(6):1754-1762. doi: 10.1021/acs.bioconjchem.9b00291. [DOI] [PubMed] [Google Scholar]
- 74.Ali HS, Boshra MS, El Meteini MS, Shafei AE, Matboli M. lncRNA-RP11-156p1.3, novel diagnostic and therapeutic targeting via CRISPR/Cas9 editing in hepatocellular carcinoma. Genomics. 2020 Sep;112(5):3306-3314. doi: 10.1016/j.ygeno.2020.06.020. [DOI] [PubMed] [Google Scholar]
- 75.Song CQ, Li Y, Mou H, et al. Genome-wide CRISPR screen identifies regulators of mitogen-activated protein kinase as suppressors of liver tumors in mice. Gastroenterology. 2017;152(5):1161-1173.e1. doi: 10.1053/j.gastro.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wei L, Lee D, Law CT, et al. Genome-wide CRISPR/Cas9 library screening identified PHGDH as a critical driver for Sorafenib resistance in HCC. Nat Commun. 2019;10(1):4681. doi: 10.1038/s41467-019-12606-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nelles DA, Fang MY, Aigner S, Yeo GW. Applications of Cas9 as an RNA-programmed RNA-binding protein. Bioessays. 2015;37(7):732-739. doi: 10.1002/bies.201500001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nelles DA, Fang MY, O’Connell MR, et al. Programmable RNA tracking in live cells with CRISPR/Cas9. Cell. 2016;165(2):488-496. doi: 10.1016/j.cell.2016.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Batra R, Nelles DA, Pirie E, et al. Elimination of toxic microsatellite repeat expansion RNA by RNA-targeting Cas9. Cell. 2017;170(5):899-912. e10. doi: 10.1016/j.cell.2017.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sheridan C. CRISPR therapeutics push into human testing. Nat Biotechnol. 2017;35(1):3-5. doi: 10.1038/nbt0117-3. [DOI] [PubMed] [Google Scholar]
- 81.Eyquem J, Mansilla-Soto J, Giavridis T, et al. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature. 2017;543(7643):113-117. doi: 10.1038/nature21405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jensen MC, Riddell SR. Designing chimeric antigen receptors to effectively and safely target tumors. Curr Opin Immunol. 2015;33:9-15. doi: 10.1016/j.coi.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stadtmauer EA, Fraietta JA, Davis MM, et al. CRISPR-engineered T cells in patients with refractory cancer. Science. 2020;367(6481):eaba7365. doi: 10.1126/science.aba7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ren J, Liu X, Fang C, Jiang S, June CH, Zhao Y. Multiplex genome editing to generate universal CAR T cells resistant to PD1 inhibition. Clin Cancer Res. 2017;23(9):2255-2266. doi: 10.1158/1078-0432.CCR-16-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang X, Rivière I. Clinical manufacturing of CAR T cells: foundation of a promising therapy. Mol Ther Oncolytics. 2016;3:16015. doi: 10.1038/mto.2016.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wright AV, Nuñez JK, Doudna JA. Biology and applications of CRISPR systems: harnessing nature’s toolbox for genome engineering. Cell. 2016;164(1-2):29-44. doi: 10.1016/j.cell.2015.12.035. [DOI] [PubMed] [Google Scholar]
- 87.Yang YC, Chen YH, Kao JH, et al. Permanent inactivation of HBV genomes by CRISPR/Cas9-mediated non-cleavage base editing. Mol Ther Nucleic Acids. 2020;20:480-490. doi: 10.1016/j.omtn.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang H, Lu H, Lei YS, et al. Development of a self-restricting CRISPR-Cas9 system to reduce off-target effects. Mol Ther Methods Clin Dev. 2020;18:390-401. doi: 10.1016/j.omtm.2020.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Maji B, Gangopadhyay SA, Lee M, et al. A high-throughput platform to identify small-molecule inhibitors of CRISPR-Cas9. Cell. 2019;177(4):1067-1079. e19. doi: 10.1016/j.cell.2019.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]