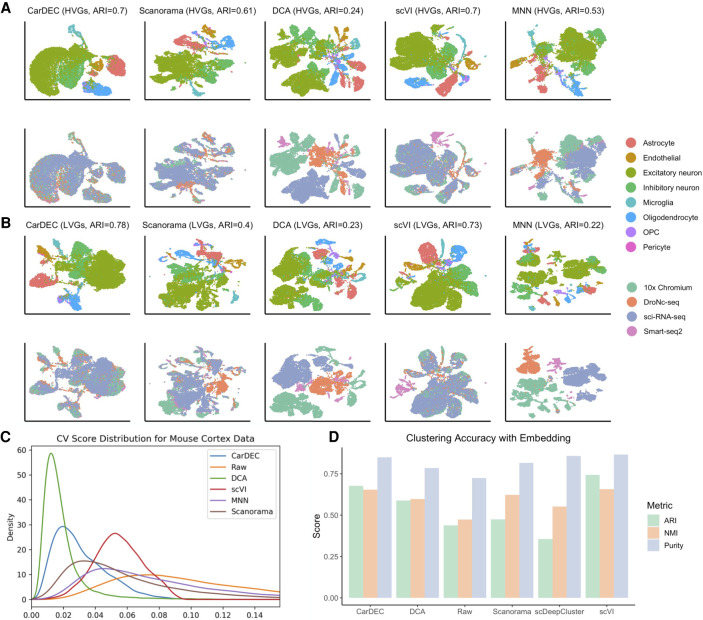
Figure 4.
Comparison of different methods on the mouse cortex data set. (A) UMAP embedding computed from the denoised HVG counts for each method. The top row was colored by cell type; the bottom row was colored by batch. Cells were also clustered with Louvain's algorithm, and the resultant ARI is provided. (B) UMAP embedding computed from the denoised LVG counts for each method. Figure legends are the same as those in A. (C) Density plot of genewise CV among batch centroids. Density plots are provided for HVGs and LVGs separately. (D) Clustering accuracy metrics obtained using the embedding-based methods to cluster the data, rather than running Louvain on the full gene expression space. Results for “Raw” were obtained by using Louvain's algorithm on the original HVG counts and are provided as a baseline against which embedding-based clustering results may be compared.