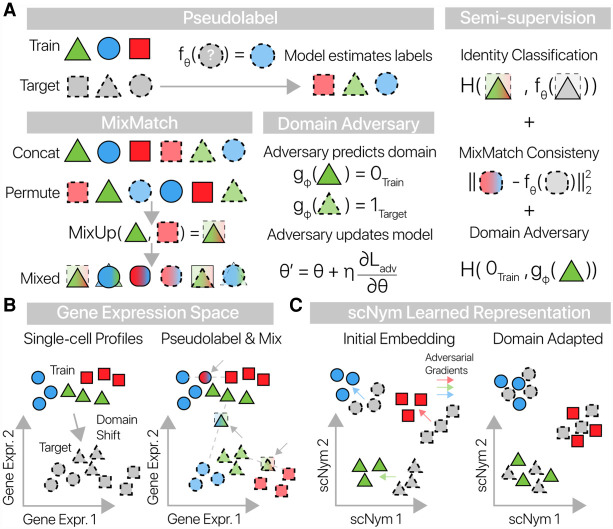
Figure 1.
scNym combines semisupervised and adversarial training to learn performant single-cell classifiers. (A) scNym takes advantage of target data during training by estimating “pseudolabels” for each target data point using model predictions. Training and target cell profiles and their labels are then augmented using weighted averages in the MixMatch procedure. An adversary is also trained to discriminate training and target observations. We train model parameters using a combination of supervised classification, interpolation consistency, and adversarial objectives. Here, we use H(·,·) to represent the cross-entropy function. (B) Training and target cell profiles are separated by a domain shift in gene expression space. scNym pseudolabels target profiles and generates mixed cell profiles (arrows) by randomly pairing cells. Mixed profiles form a bridge between training and target data sets. (C) scNym models learn a discriminative representation of cell state in a hidden embedding layer. Train and target cell profiles initially segregate in this representation. During training, adversarial gradients (colored arrows) encourage cells of the same type to mix in the scNym embedding.