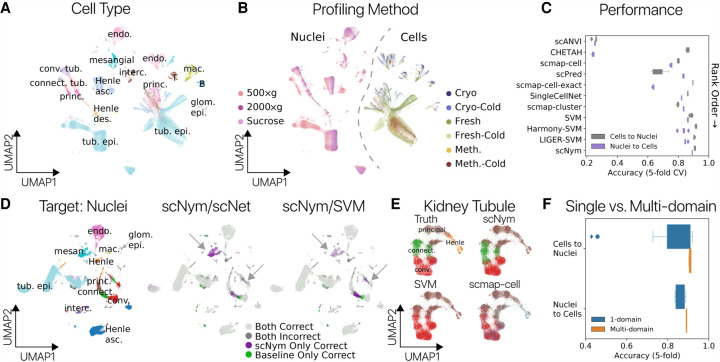
Figure 4.
Multidomain training improves cross-technology annotation transfer in the mouse kidney. Cell type (A) and sequencing protocol annotations (B) in a UMAP projection of single-cell and nucleus RNA-seq profiles from the mouse kidney (Denisenko et al. 2020). Each protocol represents a unique training domain that captures technical variation. (C) Performance of scNym and baseline approaches on single-cell–to–nucleus and single-nucleus–to–cell annotation transfer. Methods are rank ordered by performance across tasks. scNym is superior to each baseline method on at least one task (Wilcoxon rank sums, P < 0.05). (D) Single-nucleus target data labeled with true cell types (left) or the relative accuracy of scNym and baseline methods (right) for the single-cell–to–single-nucleus task. scNym achieves more accurate labeling of mesangial cells and tubule cell types (arrows). (E) Kidney tubule cells from D visualized independently with true and predicted labels. scNym offers the closest match to true annotations. All methods make notable errors on this difficult task. (F) Comparison of scNym performance when trained on individual training data sets (1-domain) versus multidomain training across all available data sets. We found that multidomain training improves performance on both the cells-to-nuclei and nuclei-to-cells transfer tasks (Wilcoxon rank sums, P = 0.073 and P < 0.01, respectively).