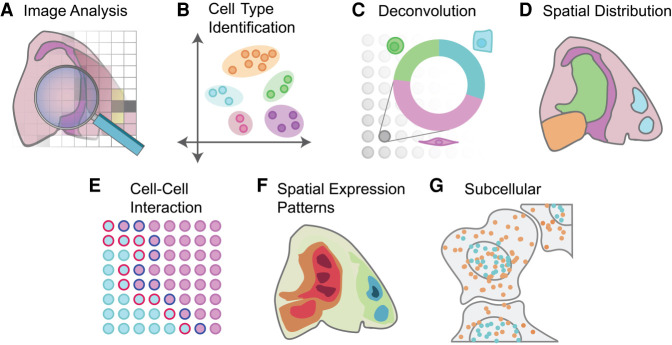
Figure 3.
Overview of spatial transcriptomics analysis methods. A variety of analyses can be performed on spatial transcriptomics data. (A) Analysis can be performed on the image itself, ranging from early tasks such as cell segmentation to support of subcellular analysis through cell shape and size classification. (B) Cell types can be identified through clustering and annotation. Additional integration with external scRNA-seq data or deconvolution of spatial units that cover multiple cells (C) can be performed to fine-tune cell type mapping. (D) The spatial distribution of cell types and the underlying cell-to-cell communication (E) can be computed. (F) Spatial expression patterns are identified and visualized based on information of gene expression and spatial coordinates. (G) Data at subcellular resolution can be used to identify spatial and temporal dynamics of transcripts within a single cell.