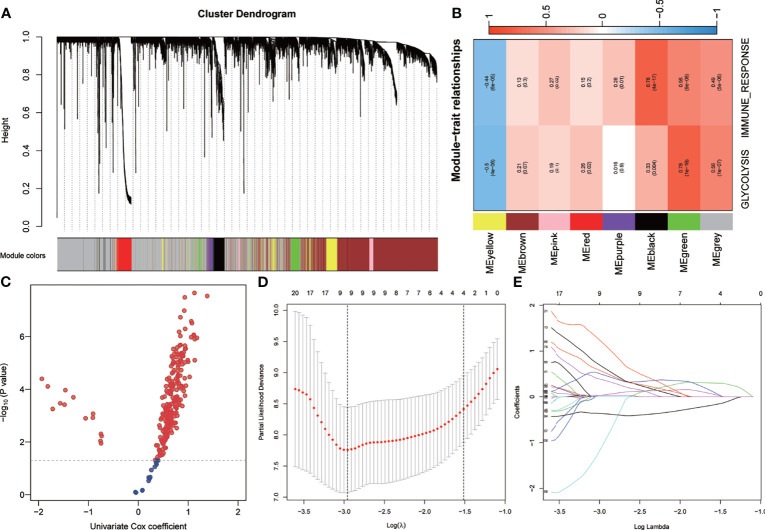
Figure 3.
Establishment of genetic signatures related to glycolysis and immune response. (A) The variance of the expression value of each gene in the training set was calculated, and then the top 6,000 genes in the variance ranking were used to construct the WGCNA network. Seven non-gray modules were identified. (B) The green module depicting the highest correlation (r > 0.5, p < 0.0001) was considered the most correlated with glycolysis and immune response. (C) Using univariate Cox regression analysis, 238 prognostic-related candidate genes were extracted from the key genes from the green module. (D, E) The LASSO Cox regression model was used to identify the most robust biomarkers, and multivariate Cox stepwise regression was applied to construct a prognostic model. LASSO, minimum absolute shrinkage and selection operator; WGCNA, weighted gene co-expression network analysis.