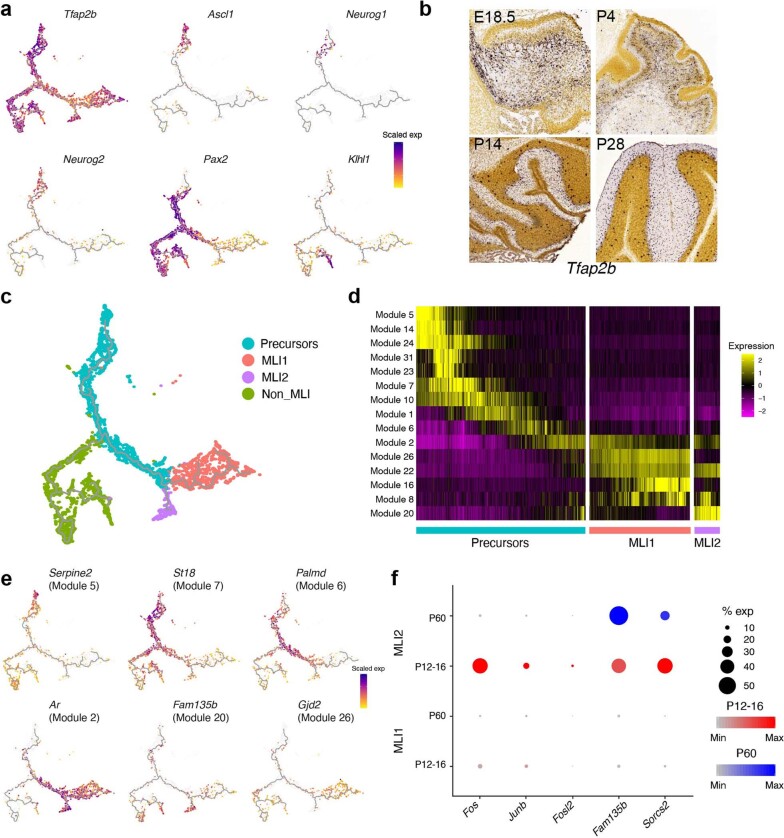
Extended Data Fig. 6. Developmental trajectory analysis of MLI1 and MLI2 neurons.
a, UMAP visualizations showing expression of canonical marker genes of cerebellar interneuron development (Tfap2b, Ascl1, Neurog1, Neurog2 and Pax2), and of mature PLIs (Klhl1). b, Allen Developing Mouse Brain Atlas expression staining of Tfap2b across four developmental time points, showing that MLI progenitors begin to enter the molecular layer around P14. c, Interneuron developmental trajectory coloured by annotated clusters. d, Heat map showing the loading of Monocle3-defined gene modules across undifferentiated interneuron precursors, MLI1 and MLI2. Cells are ordered in each block by pseudotime rank. e, UMAP visualizations showing expression of selected top-loading genes from the indicated Monocle3-defined modules. f, Dot plot showing the transient, MLI2-specific expression of three activity-regulated genes (Fos, Junb and Fosl2) and two genes whose expression persists in adulthood (Fam135b and Sorcs2).