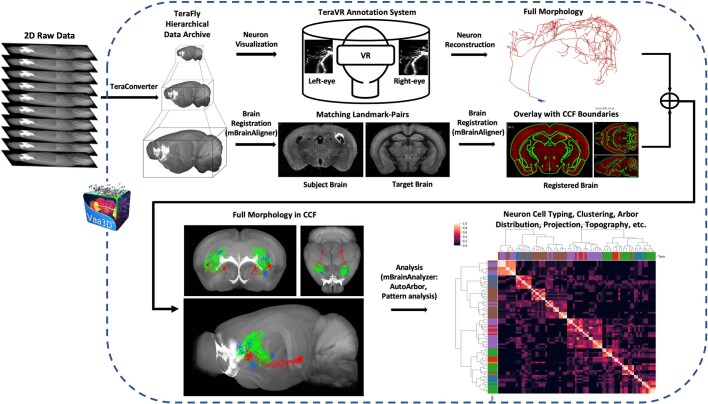
Extended Data Fig. 3. Platform and workflow of the brain-wide complete morphology imaging, reconstruction, registration and analysis pipeline.
Each fMOST dataset is first converted to a multi-level navigable TeraFly dataset using TeraConverter, the data formatting tool in the Vaa3D-TeraFly program50, which allows smooth handling of terabyte-scale datasets. Neuron visualization and reconstruction is then carried out on the TeraFly files. A series of tools, especially those based on the “Virtual Finger” method65, were developed within Vaa3D to facilitate semi-automated and manual reconstruction. Further, a virtual reality (VR) environment created within Vaa3D, TeraVR, significantly enhances a user’s ability to see the 3D relationships among intertwined axonal segments, improving precision and efficiency of reconstruction51. Annotators work in the TeraVR annotation system to reconstruct the full morphology of each neuron. After quality control (QC) and manual correction, Vaa3D’s deformable model is used to automatically fit the tracing to the center of fluorescent signals. The final reconstructed morphology is completed as a single tree without breaks, loops, or trifurcations. All these data processing, reconstruction, and workflow control processes are managed using a specific software system for massive scale data production. In parallel, each fMOST dataset is registered to CCFv3 using mBrainAligner, which uses both CLM (Coherent-Landmark-Matching) and LQW (Little-Quick-Warp) modules in brain alignment. Following registration of the whole-brain image dataset to CCFv3, all the reconstructed morphologies from the same brain are also registered for subsequent visualization and quantitative analysis. Registration to CCFv3 enables digital anatomical delineation and spatial quantification of each reconstructed morphology and its compartments (e.g., soma, dendrites, axon arbors). Since neurons are reconstructed from different brains, co-registration to the CCFv3 allows them to be compared and analyzed using a unified framework, mBrainAnalyzer, which automatically detects the arbors of each neuron followed by mapping of these dendritic and axonal arbors onto the standardized CCFv3 space. Morphological features such as length, depth, area, etc., at the whole neuron level are also computed for each arbor-domain for analysis.