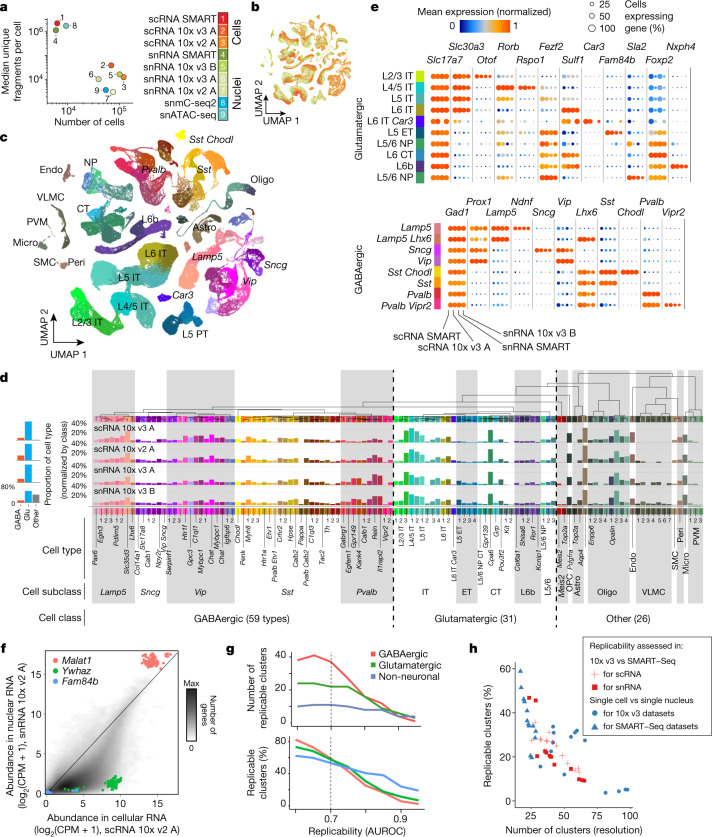
Fig. 1. Multi-platform transcriptomic taxonomy of the cell types in the MOp.
a, Key attributes of nine single-cell transcriptomic and epigenomic datasets from the mouse MOp. b, c, Two-dimensional projection (uniform manifold approximation and projection (UMAP)40) of cells and nuclei based on integrated analysis of seven transcriptomic (scRNA-seq and snRNA-seq) datasets. Cells and nuclei are coloured by dataset (b) using the colours shown in a, or by cell type (c). Non-neuronal cell types are depleted owing to the sampling strategy, which enriched neurons in all datasets except snRNA 10x v3 B. d, Dendrogram showing a hierarchical relationship among the consensus transcriptomic cell types and a proportion of cells of each type per dataset, normalized within major classes. Glu, glutamatergic. e, Expression of selected marker genes for excitatory (top) and inhibitory (bottom) cell classes, across four platforms. f, Differential enrichment of transcripts in single cells versus single nuclei. The long non-coding RNA Malat1 is enriched in nuclei. CPM, counts per million reads mapped. g, The number of replicable clusters across at least two of the seven scRNA-seq and snRNA-seq datasets as a function of the minimal MetaNeighbor score (AUROC). h, The trade-off between the number of clusters and replicability (the per cent of clusters with minimal MetaNeighbor replicability score). The major inhibitory neuron subclasses are Lamp5, Sncg, Vip, Sst and Pvalb. Astro, astrocyte; CT, corticothalamic; endo, endothelial; ET, extratelencephalically projecting; IT, intratelencephalically projecting; micro, microglial cell; NP, near-projecting; oligo, oligodendrocyte; OPC, oligodendrocyte precursor cell; peri, pericyte; PVM, perivascular macrophage; SMC, smooth muscle cell; VLMC, vascular leptomeningeal cell.