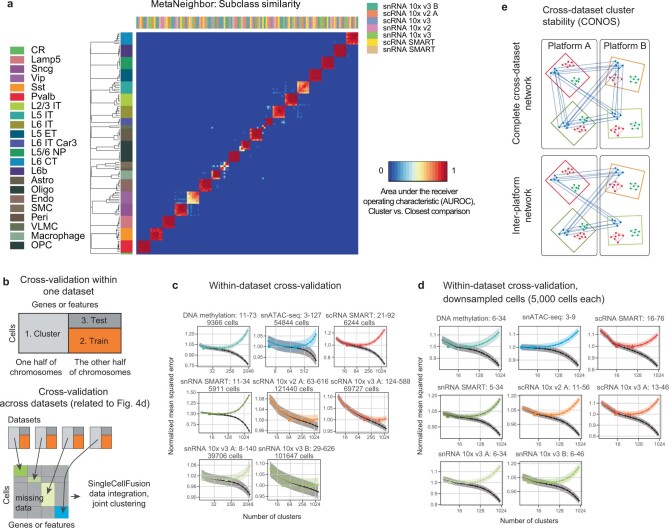
Extended Data Fig. 8. MetaNeighbor and cross-validation analysis of cluster reproducibility.
a, Heat map showing replicability scores (MetaNeighbor AUROC) at the subclass level of the independent clusterings of seven RNA-seq datasets. High AUROC indicates that the cell-type labels in one dataset can be reliably predicted based on the nearest neighbours of those cells in another dataset, together with the independent cluster analysis of that dataset. b, Scheme for within-dataset and across-dataset cross-validation. c, d Within-dataset cross-validation analysis for each dataset, either using the full set of cells (c) or using a random sample of 5,000 cells (d). In each plot, the black curve shows training error, while the coloured U-shaped curve shows the test set error, with a minimum at the cluster resolution that balances over-fitting and under-fitting. The shaded region shows the s.e.m. based on cross-validation with n = 5 data partitions. e, Transcriptomic platform consistency is assessed by cross-dataset cluster stability analysis (Conos37).