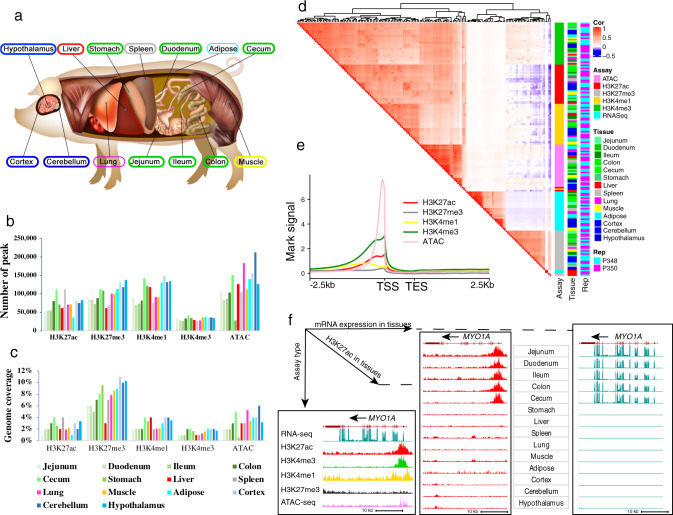
Fig. 1. Data summary of epigenomic information across tissues and marks.
a Tissues assayed by this study. b, c Average peak number and genome coverage for each epigenetic mark in each tissue. d The Pearson correlations among assays, tissues, and biological replicates (P348 and P350) based on the normalized signal in 1 kb windows stepped across the whole genome. e Average epigenetic mark signal proximal to protein-coding genes. TSS transcription start site, TES transcription end site. f Epigenetic signal at the MYO1A locus according to different assays and in different tissues. Vertical scale of UCSC tracks shows normalized signal from 0 to 200 for RNA-seq, 0 to 100 for H3K27ac and H3K4me3, and 0 to 50 for other marks and ATAC-seq.