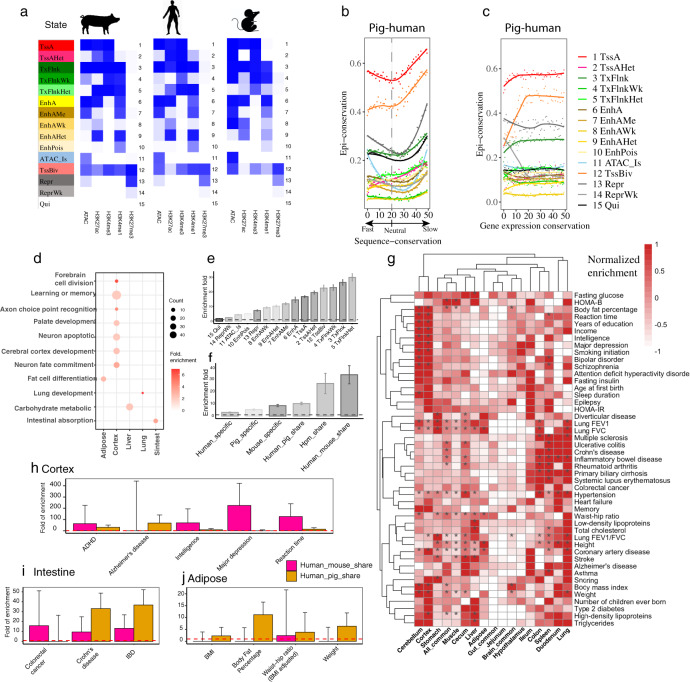
Fig. 6. Interspecies conservation of chromatin states.
a 15 chromatin states predicated in three species. The colors from white to deep blue indicate emission probabilities, ranging from 0 to 1. b Relation between sequence conservation and epigenomic conservation across six tissues. Fifty genomic regions were ordered from the fastest changing (0th), neutral (20th), and slowest changing (49th) in terms of sequence conservation (Supplementary Fig. 16d). Epigenome conservation (see Methods section) for each chromatin state within each region was calculated between pigs and humans and plotted. c Relation between expression conservation and epigenomic conservation across six tissues. Expression conservation was based on expression of 14,302 orthologous genes among the three species. Regions were ordered from the biggest difference in expression (0th), to the smallest difference (49th). d GO enrichment was based on genes proximal to (±2 kb) human-specific TssA with extreme sequence conservation (49th). Count refers to the number of genes. e Human GWAS (47 traits) signal enrichment in 15 different chromatin states across six tissues. Enrichment was the proportion of heritability divided by the proportion of SNPs in each chromatin state. Values greater than the dashed line (set at 1) indicate significant enrichment. Error bars represent standard error around the estimates of enrichment. Dashed lines and error bars are similarly formatted in sub-figures (f, h–j). f Human GWAS (47 traits) enrichment in six groups of species-specific or shared EnhA across six tissues. (hpm_share stands for human-pig-mouse shared). g GWAS enrichment of pig tissue-specific enhancer (EnhA) in humans. “*” indicates FDR < 0.05. h–j Different GWAS enrichments between human-pig and human-mouse shared strong enhancers (EnhA) in brain cortex (1799 vs. 61 enhancers), small intestine (5311 vs. 2430 enhancers), and adipose (2014 vs. 1638 enhancers), respectively. Data in Figs. e–j are available at 10.6084/m9.figshare.16531197.v1.