Fig. 2. SaPI TerS drives efficient packaging of plasmid DNA into transducing particles.
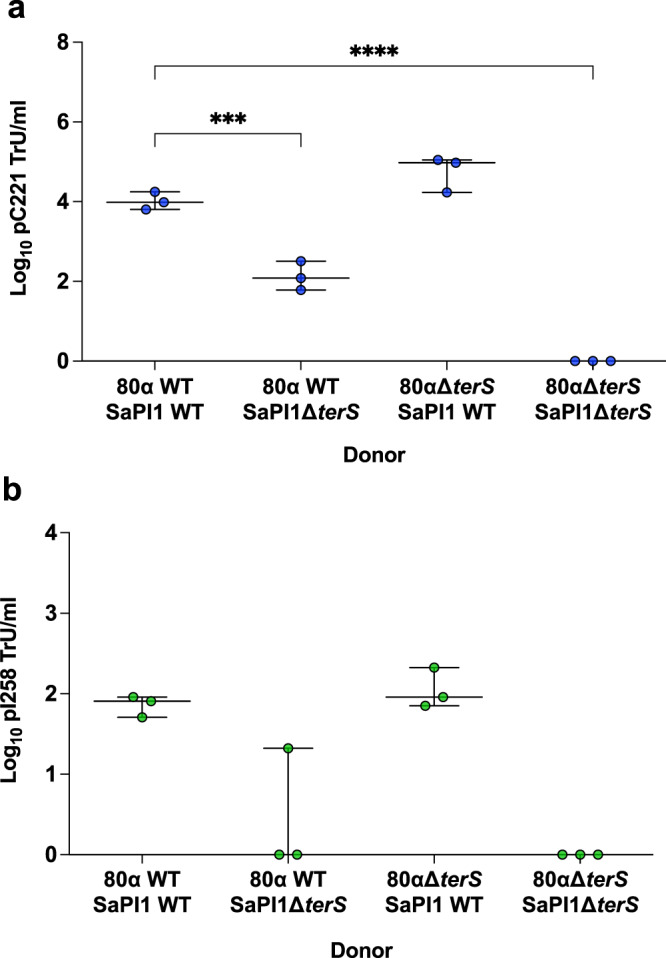
RN4220 strains lysogenic for phage 80α (WT or mutant in small terminase) and SaPI1 (WT or mutant in small terminase), carrying plasmid a pC221 (4.6 kb; blue circles) or b or pI258 (29 kb; green circles) were induced with mitomycin C to produce lysates. Log10 transductants (TrU) per ml of transducing lysate were determined for each plasmid in an RN4220 recipient. All data is the result of three independent experiments (n = 3). Data for each donor strain are represented as boxplots where the middle line is the median, the lower and upper hinges correspond to the 25th and 75th percentiles, and the whiskers extend from the minimum to maximum values, with all individual data points shown as coloured circles. For a, a one-way ANOVA with Tukey’s multiple comparisons test compared mean differences between each strain and the 80α WT SaPI1 WT control. Asterisks denote significant adjusted p values: ***p = 0.0003, ****p < 0.0001. For b, a Kruskal–Wallis (one-way ANOVA on ranks) with Dunn’s multiple comparisons test compared mean rank values between each strain and the 80α WT SaPI1 WT control. No statistically significant differences were observed between the groups (p > 0.05).
