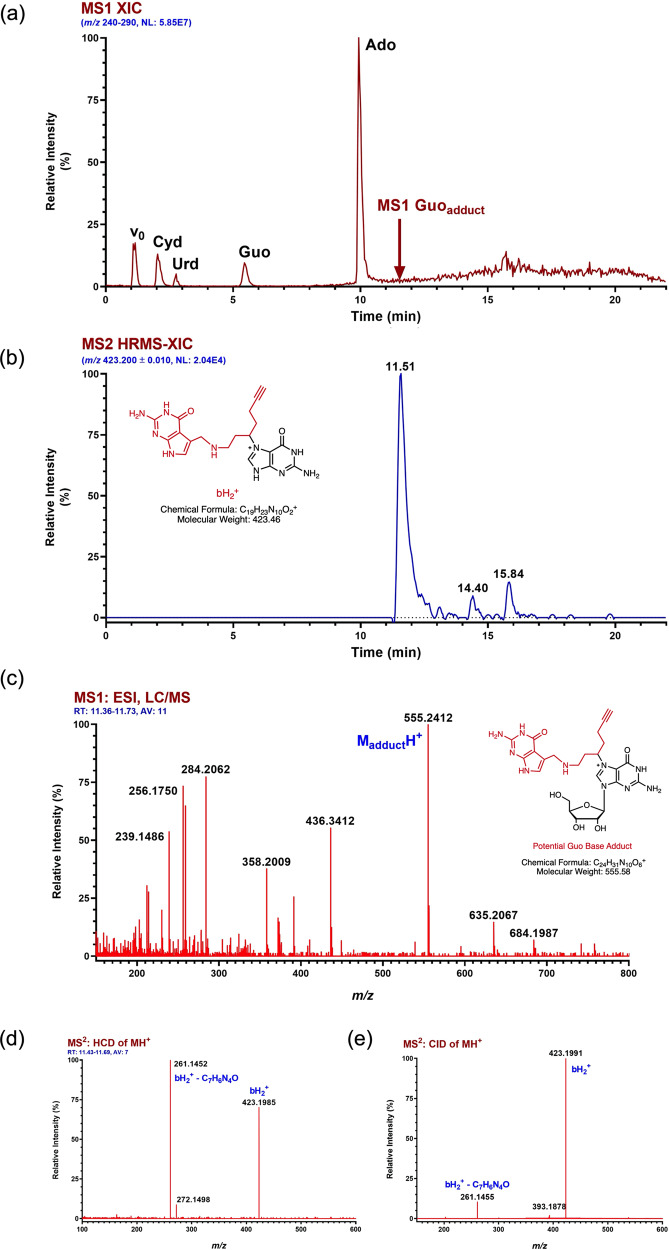
Fig. 3. Probe 11 selectively crosslinks to guanosine bases in the PreQ1 aptamer.
High-resolution accurate mass (HRAM) and positive ion LC/MS/MS analysis of the nucleoside digest resulting from adducted Ss-preQ1-RS (RNA 35-mer). a MS1 extracted ion chromatogram (XIC) for m/z 240–290 showing the response for unmodified RNA nucleosides. HPLC void volume is indicated by v0 and the elution time of the major RNA probe adduct is indicated by a red arrow. b MS2 (CID of Guo-adduct MH+) HRAM XIC for bH2+ , the ion indicating that adduct formation occurs on the base. Extracted ion mass tolerance is ±10 mDa. The ion structure represents one of several possibilities for adduct and charge location. c Averaged and background-subtracted high-resolution MS1 spectrum for the major Guo-adduct located through HRAM XIC analysis of the theoretical MH+ (m/z 555.2423) for potential adducts. A plausible structure of the adduct is illustrated. d Full scan high-resolution MS/MS spectrum for HCD of MH+ of the Guo-adduct eluting at 11.51 min in a & b. e Full-range high-resolution MS/MS spectrum for CID of the same Guo-adduct eluting at 11.51 min in a & b.