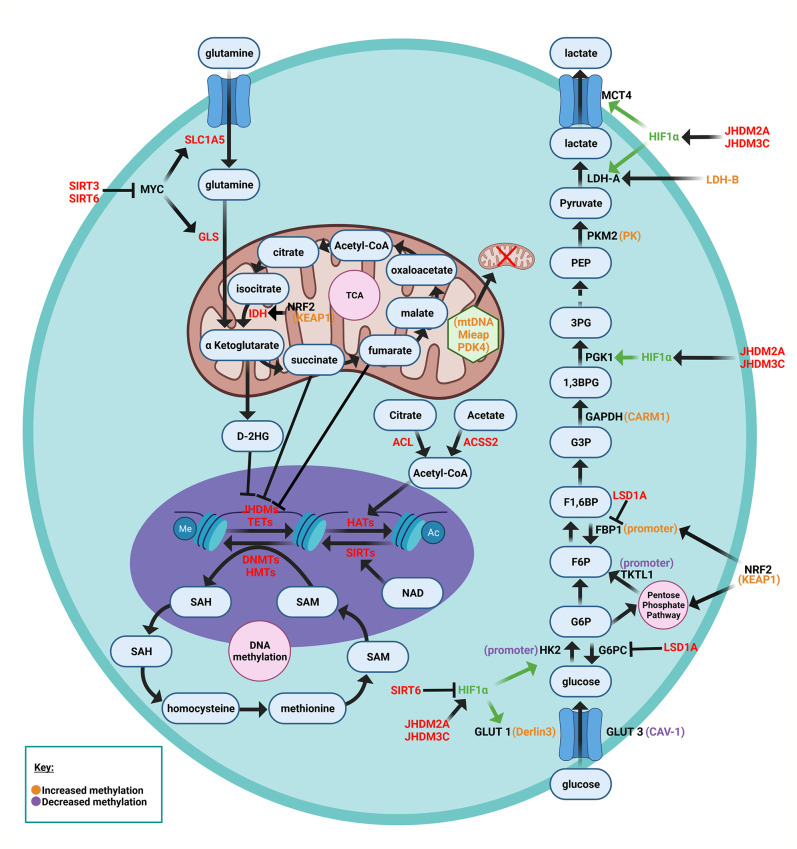
Figure 2.
Schematic representation of metabolic nodes governed by epigenetic modification. Metabolic enzymes and nutrient transporters are governed by modification by DNA methylation and histone modifications. Bidirectional feedback occurs as the metabolic substrates generated in turn regulate epigenetic modifications. α-ketoglutarate is a co-substrate for JHDM (The Jumonji-domain histone demethylase) histone demethylase and the TET family methylcytosine dioxygenases, thereby governing demethylation of histone and other proteins, and DNA. Fumarate, succinate and 2-HG compete with a ketoglutarate. HIF1α (hypoxia inducible factor 1α), KEAP1 (Kelch-like ECH-associated protein 1), NRF2 (nuclear factor erythroid 2-related factor 2), FBP1 (fructose-1,6-bisphosphate isoform 1), TKTL1 (transketolase (TKT) like-1 gene), SAM (S-adenosyl methionine), TCA (tricarboxylic acid), The abundance of GLUT3 and GLUT1 are regulated by CAV-1 and Derlin3 respectively.