Figure 5.
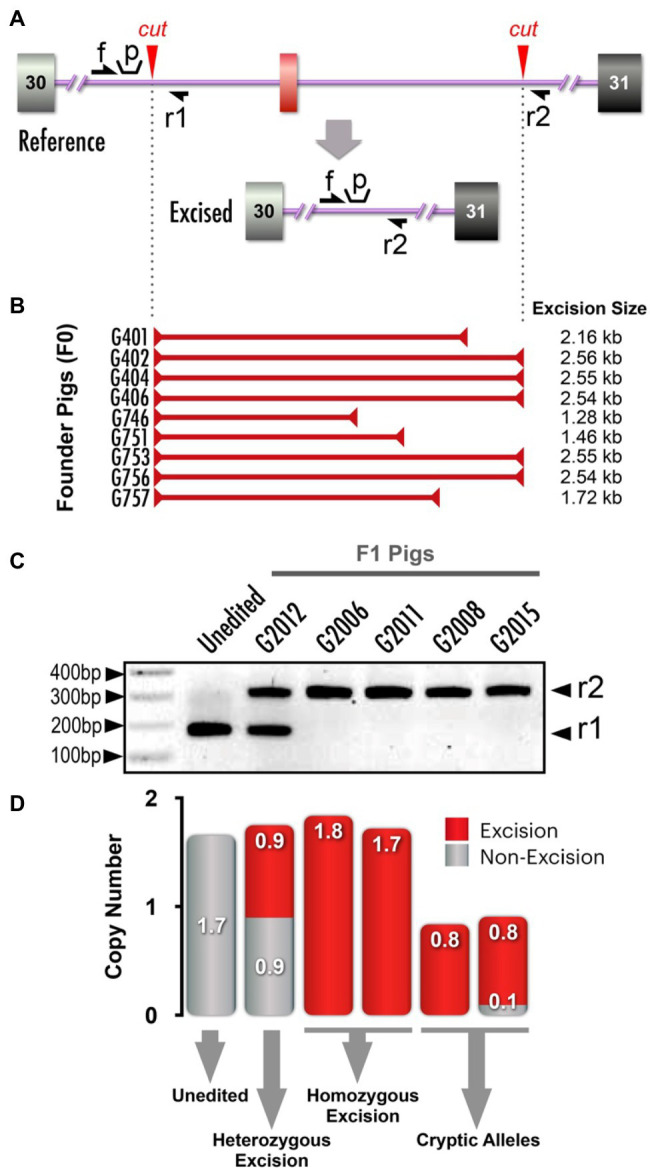
NF1 splicing variants (NF1 a31 excision) and copy number variance (CNV) analysis. (A) Schematic depicts alterations made within flanking regions of NF1 exon a31. Red arrowheads: Cas9 cut sites, single-headed black arrows (f, r1, r2): primer annealing sites for competitive and dPCR (distinct primer sets, see Supplementary Table 2 for primer sequences), “p” indicates dPCR probe annealing site. (B) PCR analysis identified 10 out of 35 founder pigs as carrying the a31 excision. The breakpoints of each excision allele are mapped to scale, depicting the diversity of recovered excision alleles (only two were identical). Sanger sequencing identified excisions ranging from 1.28kb to the expected 2.56kb. (C) PCR amplicon products from F1 pigs using primers indicated in (A). See Supplementary Figure 2 for complete gel electrophoresis image. F1 pigs from founders with identical excision alleles revealed the presence of excision and non-excision alleles. Amplification of excision alleles and non-excision alleles yielded products of different sizes, indicated with black arrowheads on right (r1 and r2). Unedited and heterozygote (non-cryptic) are shown, along with four pigs in which only the excision allele was detected (i.e., a non-excision allele was not detected). (D) CNV quantification determined separately for excision and non-excision dPCR (red and grey, respectively) and plotted together. Note that for some F1 pigs only a single copy was detected in both PCRs, indicating that these animals carry cryptic mutations that cannot be readily detected.
